mLN EYFP+ all timepoints
A.DeMartin
2025-04-29
Last updated: 2025-07-15
Checks: 6 1
Knit directory: LNdevMouse24.2/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250625) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 6788aae. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Untracked files:
Untracked: analysis/mLNEYFPpos.Rmd
Untracked: analysis/tradeSeq.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
load packages
library(ExploreSCdataSeurat3)
library(runSeurat3)
library(Seurat)
library(ggpubr)
library(pheatmap)
library(SingleCellExperiment)
library(dplyr)
library(tidyverse)
library(viridis)
library(muscat)
library(circlize)
library(destiny)
library(scater)
library(metap)
library(multtest)
library(clusterProfiler)
library(org.Hs.eg.db)
library(msigdbr)
library(enrichplot)
library(DOSE)
library(grid)
library(gridExtra)
library(ggupset)
library(VennDiagram)
library(NCmisc)
library(slingshot)
library(RColorBrewer)
library(here)preprocessing
load object all
basedir <- here()
fileNam <- paste0(basedir, "/data/LNmLToRev_allmerged_seurat.rds")
seuratM <- readRDS(fileNam)
table(seuratM$dataset)
380131_02-2_20250224_Cxcl13EYFP_P7_iLN_YFPneg 380131_03-3_20250224_Cxcl13EYFP_P7_iLN_YFPpos
5138 3906
380131_04-4_20250224_Cxcl13EYFP_P7_mLN_YFPpos 380131_05-5_20250224_Cxcl13EYFP_P7_mLN_YFPneg
8203 6337
380131_06-6_20250225_Cxcl13EYFP_E18_iLN_fib 380131_07-7_20250225_Cxcl13EYFP_E18_mLN_YFPpos
5362 5438
380131_08-8_20250225_Cxcl13EYFP_E18_mLN_YFPneg 380131_11-11_20250305_Mu_Cxcl13EYFP_Adult_pLN_FRC
5449 5344
380131_12-12_20250305_Mu_Cxcl13EYFP_Adult_mLN_FRC 382581_01-1_20250311_Mu_Cxcl13EYFP_E18_iLN_fib
6746 4035
382581_02-2_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPpos 382581_03-3_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPneg
8101 14326
382581_04-4_20250319_Mu_Cxcl13EYFP_P7_iLN_YFPpos 382581_05-5_20250319_Mu_Cxcl13EYFP_P7_iLN_YFPneg
3107 5161
382581_06-6_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPpos 382581_07-7_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPneg
8557 4427
382581_08-8_20250320_Mu_Cxcl13EYFP_Adult_pLN_FRC 382581_09-9_20250320_Mu_Cxcl13EYFP_Adult_mLN_FRC
7338 3739
382581_12-12_20250402_Mu_Cxcl13EYFP_3wk_iLN_fib1 382581_13-13_20250402_Mu_Cxcl13EYFP_3wk_iLN_fib2
6689 8423
382581_14-14_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib1 382581_15-15_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib2
6477 7988 table(seuratM$RNA_snn_res.0.25)
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
26907 18102 16879 15100 12710 7758 6842 6775 5471 5456 5030 3609 2710 1779 1650 958
16 17 18 19
851 829 508 367 table(seuratM$orig.ident)
140291 subset EYFP+
seuratSub <- subset(seuratM, Rosa26eyfp.Rosa26eyfp>0)
eyfpPos <- colnames(seuratSub)
seuratM$EYFP <-"neg"
seuratM$EYFP[which(colnames(seuratM)%in%eyfpPos)] <- "pos"
table(seuratM$dataset, seuratM$EYFP)
seuratEYFP <- subset(seuratM, EYFP == "pos")
table(seuratEYFP$orig.ident)
## rerun seurat
seuratEYFP <- NormalizeData (object = seuratEYFP)
seuratEYFP <- FindVariableFeatures(object = seuratEYFP)
seuratEYFP <- ScaleData(object = seuratEYFP, verbose = TRUE)
seuratEYFP <- RunPCA(object=seuratEYFP, npcs = 30, verbose = FALSE)
seuratEYFP <- RunTSNE(object=seuratEYFP, reduction="pca", dims = 1:20)
seuratEYFP <- RunUMAP(object=seuratEYFP, reduction="pca", dims = 1:20)
seuratEYFP <- FindNeighbors(object = seuratEYFP, reduction = "pca", dims= 1:20)
res <- c(0.25, 0.6, 0.8, 0.4)
for (i in 1:length(res)) {
seuratEYFP <- FindClusters(object = seuratEYFP, resolution = res[i], random.seed = 1234)
}## save object
saveRDS(seuratEYFP, file=paste0(basedir,"/data/LNmLToRev_EYFPonly_seurat.rds"))load object EYFP+
fileNam <- paste0(basedir, "/data/LNmLToRev_EYFPonly_seurat.rds")
seuratEYFP <- readRDS(fileNam)
table(seuratEYFP$dataset)
380131_02-2_20250224_Cxcl13EYFP_P7_iLN_YFPneg 380131_03-3_20250224_Cxcl13EYFP_P7_iLN_YFPpos
73 3026
380131_04-4_20250224_Cxcl13EYFP_P7_mLN_YFPpos 380131_05-5_20250224_Cxcl13EYFP_P7_mLN_YFPneg
4780 24
380131_06-6_20250225_Cxcl13EYFP_E18_iLN_fib 380131_07-7_20250225_Cxcl13EYFP_E18_mLN_YFPpos
165 3604
380131_08-8_20250225_Cxcl13EYFP_E18_mLN_YFPneg 380131_11-11_20250305_Mu_Cxcl13EYFP_Adult_pLN_FRC
13 3424
380131_12-12_20250305_Mu_Cxcl13EYFP_Adult_mLN_FRC 382581_01-1_20250311_Mu_Cxcl13EYFP_E18_iLN_fib
3951 1095
382581_02-2_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPpos 382581_03-3_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPneg
5457 29
382581_04-4_20250319_Mu_Cxcl13EYFP_P7_iLN_YFPpos 382581_05-5_20250319_Mu_Cxcl13EYFP_P7_iLN_YFPneg
2689 99
382581_06-6_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPpos 382581_07-7_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPneg
6111 23
382581_08-8_20250320_Mu_Cxcl13EYFP_Adult_pLN_FRC 382581_09-9_20250320_Mu_Cxcl13EYFP_Adult_mLN_FRC
4279 2829
382581_12-12_20250402_Mu_Cxcl13EYFP_3wk_iLN_fib1 382581_13-13_20250402_Mu_Cxcl13EYFP_3wk_iLN_fib2
3261 3590
382581_14-14_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib1 382581_15-15_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib2
3809 5297 table(seuratEYFP$RNA_snn_res.0.25)
0 1 2 3 4 5 6 7 8 9 10 11 12 13
13255 7957 7189 5621 4892 4822 4378 2353 1924 1732 1005 945 855 700 table(seuratEYFP$orig.ident)
57628 subset mLN
## subset
table(seuratEYFP$location)
seuratmLN <- subset(seuratEYFP, location == "mLN")
table(seuratmLN$orig.ident)
## rerun seurat
seuratmLN <- NormalizeData (object = seuratmLN)
seuratmLN <- FindVariableFeatures(object = seuratmLN)
seuratmLN <- ScaleData(object = seuratmLN, verbose = TRUE)
seuratmLN <- RunPCA(object=seuratmLN, npcs = 30, verbose = FALSE)
seuratmLN <- RunTSNE(object=seuratmLN, reduction="pca", dims = 1:20)
seuratmLN <- RunUMAP(object=seuratmLN, reduction="pca", dims = 1:20)
seuratmLN <- FindNeighbors(object = seuratmLN, reduction = "pca", dims= 1:20)
res <- c(0.25, 0.6, 0.8, 0.4)
for (i in 1:length(res)) {
seuratmLN <- FindClusters(object = seuratmLN, resolution = res[i], random.seed = 1234)
}### save object
saveRDS(seuratmLN, file=paste0(basedir,"/data/LNmLToRev_EYFP_mLN_seurat.rds"))load object mLN EYFP+
fileNam <- paste0(basedir, "/data/LNmLToRev_EYFP_mLN_seurat.rds")
seuratmLN <- readRDS(fileNam)
table(seuratmLN$dataset)
380131_04-4_20250224_Cxcl13EYFP_P7_mLN_YFPpos 380131_05-5_20250224_Cxcl13EYFP_P7_mLN_YFPneg
4780 24
380131_07-7_20250225_Cxcl13EYFP_E18_mLN_YFPpos 380131_08-8_20250225_Cxcl13EYFP_E18_mLN_YFPneg
3604 13
380131_12-12_20250305_Mu_Cxcl13EYFP_Adult_mLN_FRC 382581_02-2_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPpos
3951 5457
382581_03-3_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPneg 382581_06-6_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPpos
29 6111
382581_07-7_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPneg 382581_09-9_20250320_Mu_Cxcl13EYFP_Adult_mLN_FRC
23 2829
382581_14-14_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib1 382581_15-15_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib2
3809 5297 table(seuratmLN$RNA_snn_res.0.25)
0 1 2 3 4 5 6 7 8 9 10 11 12 13
8201 5773 4993 4725 3803 1887 1706 1582 866 692 638 603 388 70 table(seuratmLN$orig.ident)
35927 set color vectors
coltimepoint <- c("#440154FF", "#3B528BFF", "#21908CFF", "#5DC863FF")
names(coltimepoint) <- c("E18", "P7", "3w", "8w")
colPal <- c("#DAF7A6", "#FFC300", "#FF5733", "#C70039", "#900C3F", "#b66e8d",
"#61a4ba", "#6178ba", "#54a87f", "#25328a",
"#b6856e", "#0073C2FF", "#EFC000FF", "#868686FF", "#CD534CFF",
"#7AA6DCFF", "#003C67FF", "#8F7700FF", "#3B3B3BFF", "#A73030FF",
"#4A6990FF")[1:length(unique(seuratmLN$RNA_snn_res.0.4))]
names(colPal) <- unique(seuratmLN$RNA_snn_res.0.4)
collocation <- c("#61baba", "#ba6161")
names(collocation) <- c("iLN", "mLN")dimplot
clustering
DimPlot(seuratmLN, reduction = "umap", group.by = "RNA_snn_res.0.4",
cols = colPal, label = TRUE)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")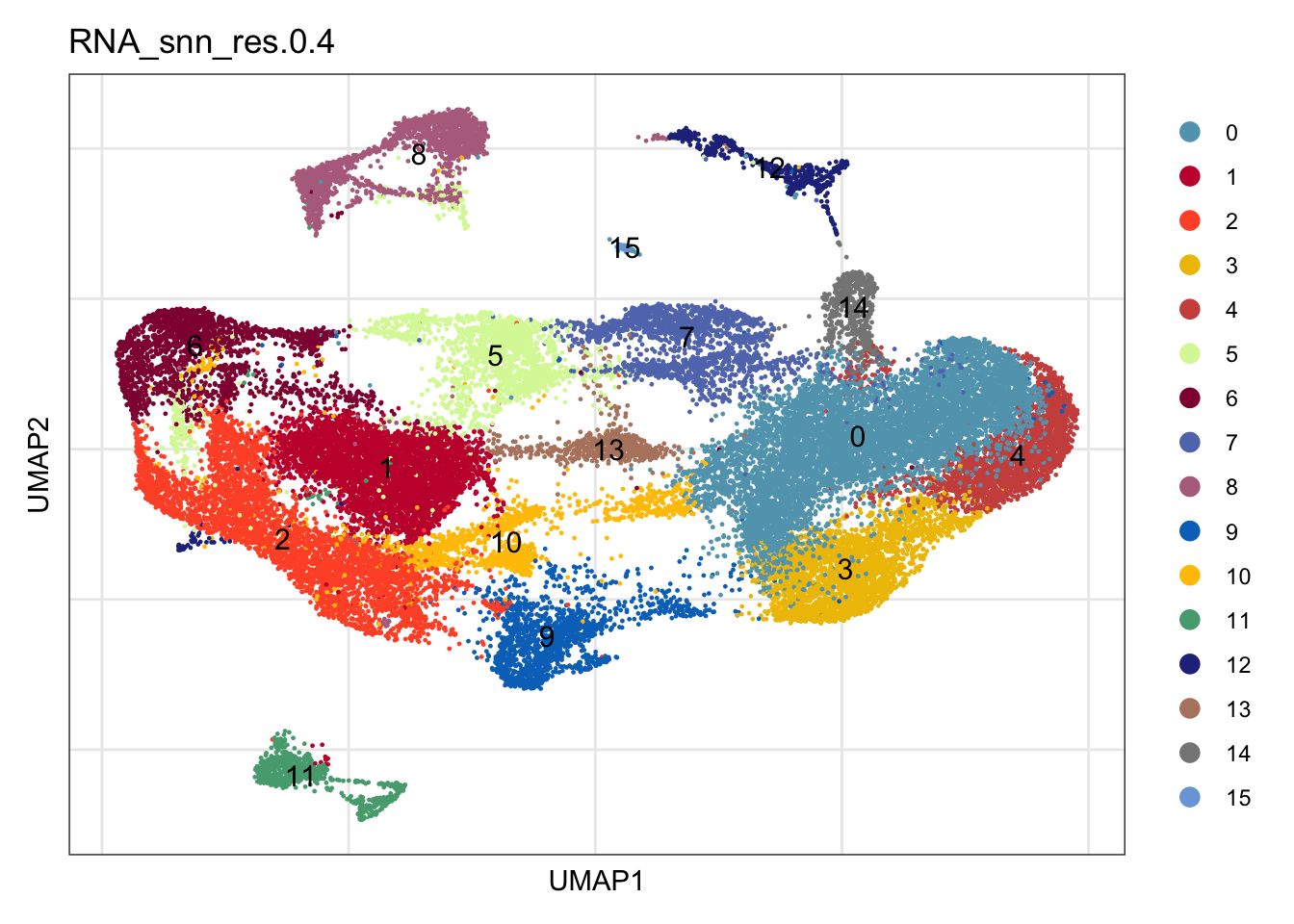
timepoint
DimPlot(seuratmLN, reduction = "umap", group.by = "timepoint",
cols = coltimepoint)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")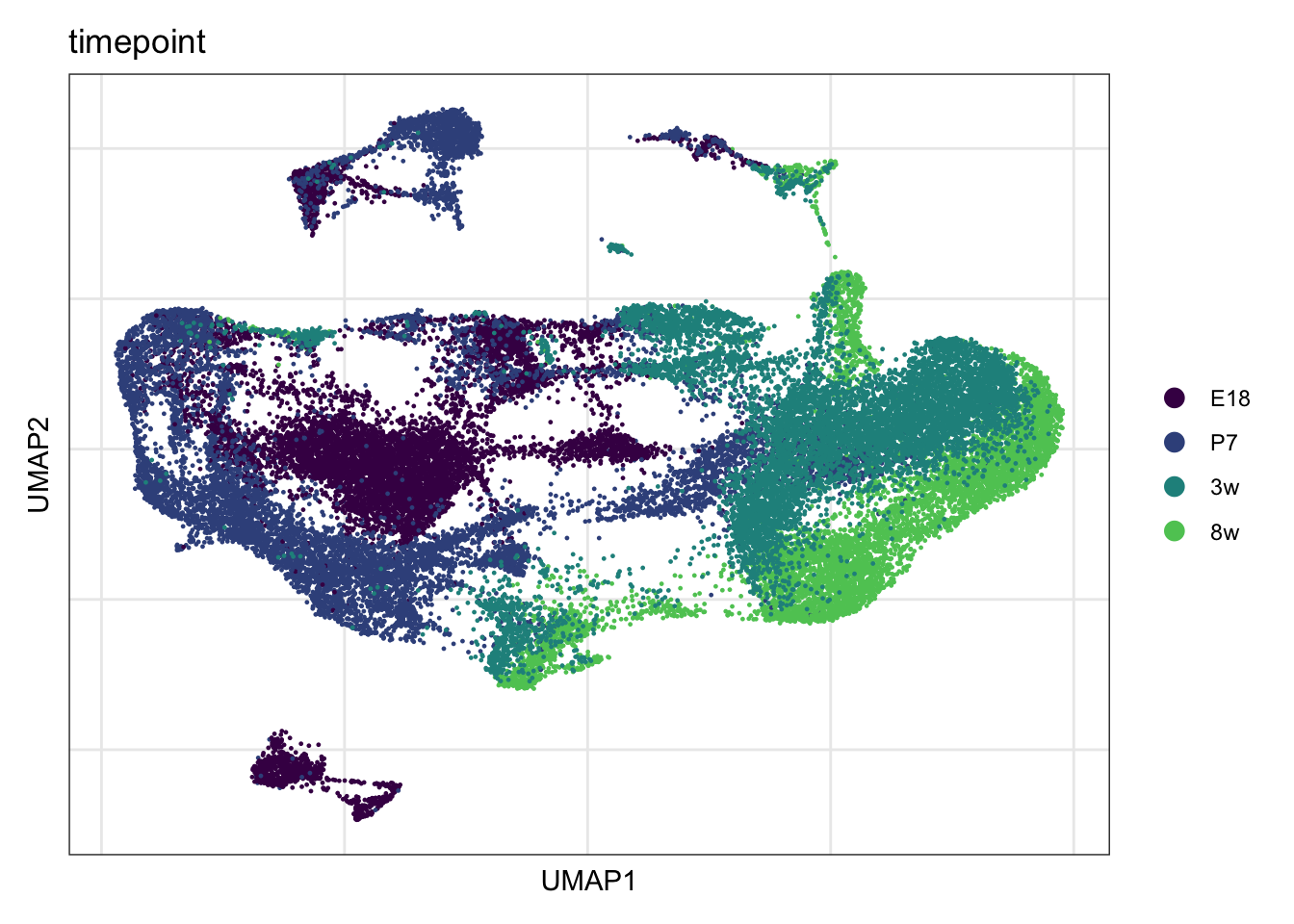
calculate cluster marker genes
## cluster marker
Idents(seuratmLN) <- seuratmLN$RNA_snn_res.0.4
markerGenes <- FindAllMarkers(seuratmLN, only.pos=T) %>%
dplyr::filter(p_val_adj < 0.01)features
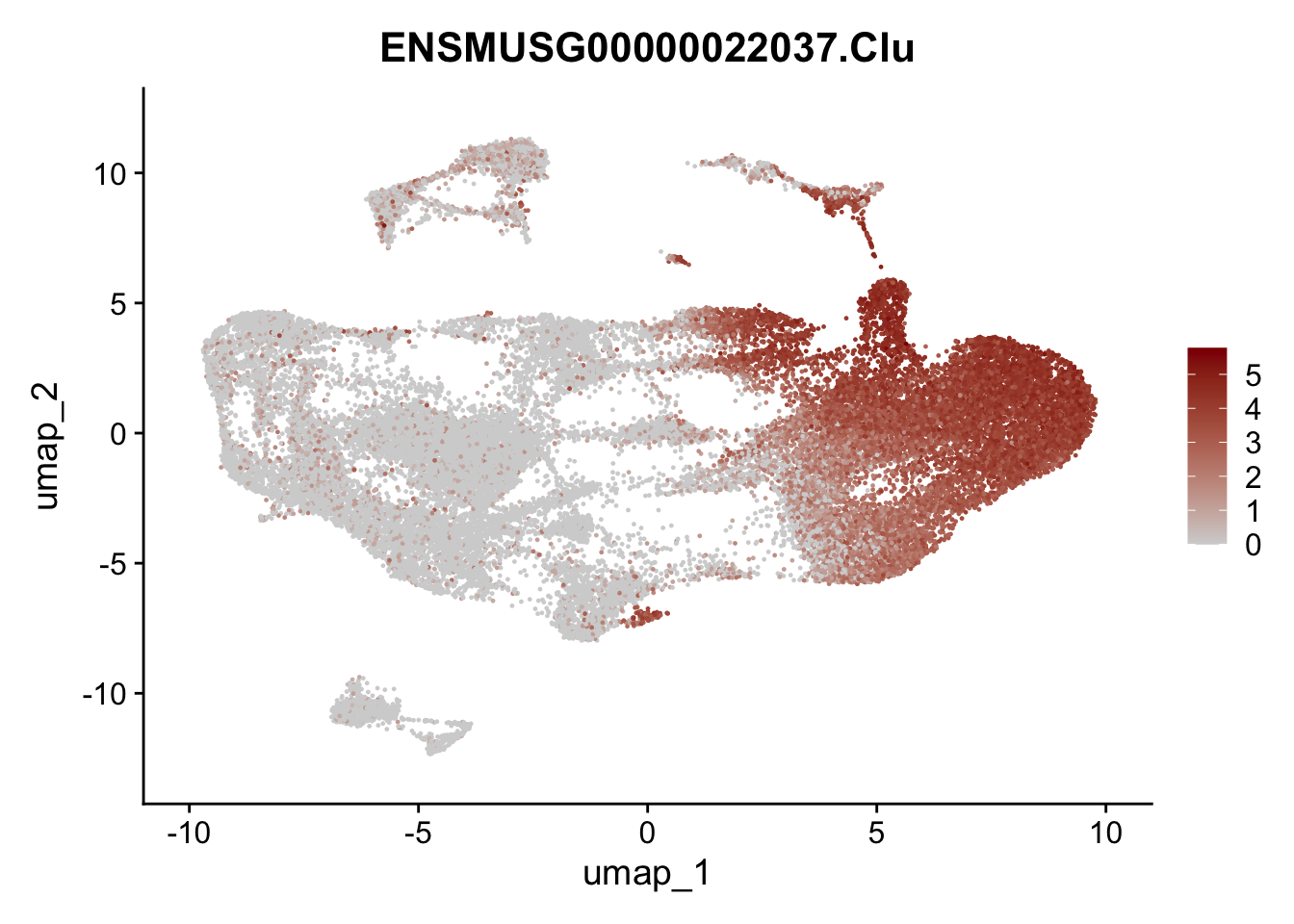
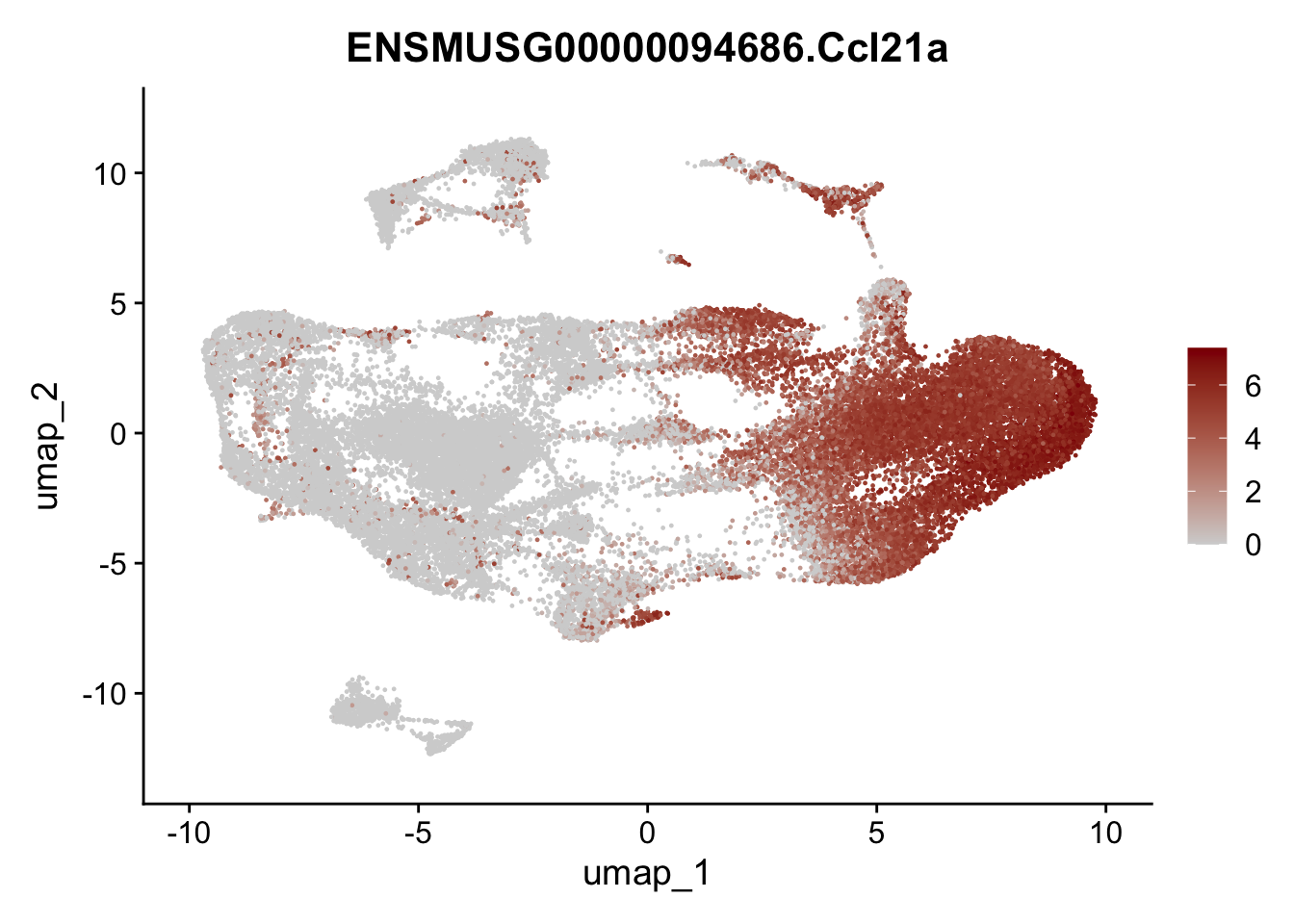
genes <- data.frame(gene=rownames(seuratmLN)) %>%
mutate(geneID=gsub("^.*\\.", "", gene))
selGenesAll <- data.frame(geneID=c("Clu", "Ccl21a", "Grem1", "Grem2","Mfap4", "Ccl19","Cd34", "Col6a1",
"Col6a2","Rosa26eyfp", "Cxcl13", "Icam1", "Vcam1")) %>%
left_join(., genes, by = "geneID")
pList <- sapply(selGenesAll$gene, function(x){
p <- FeaturePlot(seuratmLN, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = F)+
theme(legend.position="right")
plot(p)
})
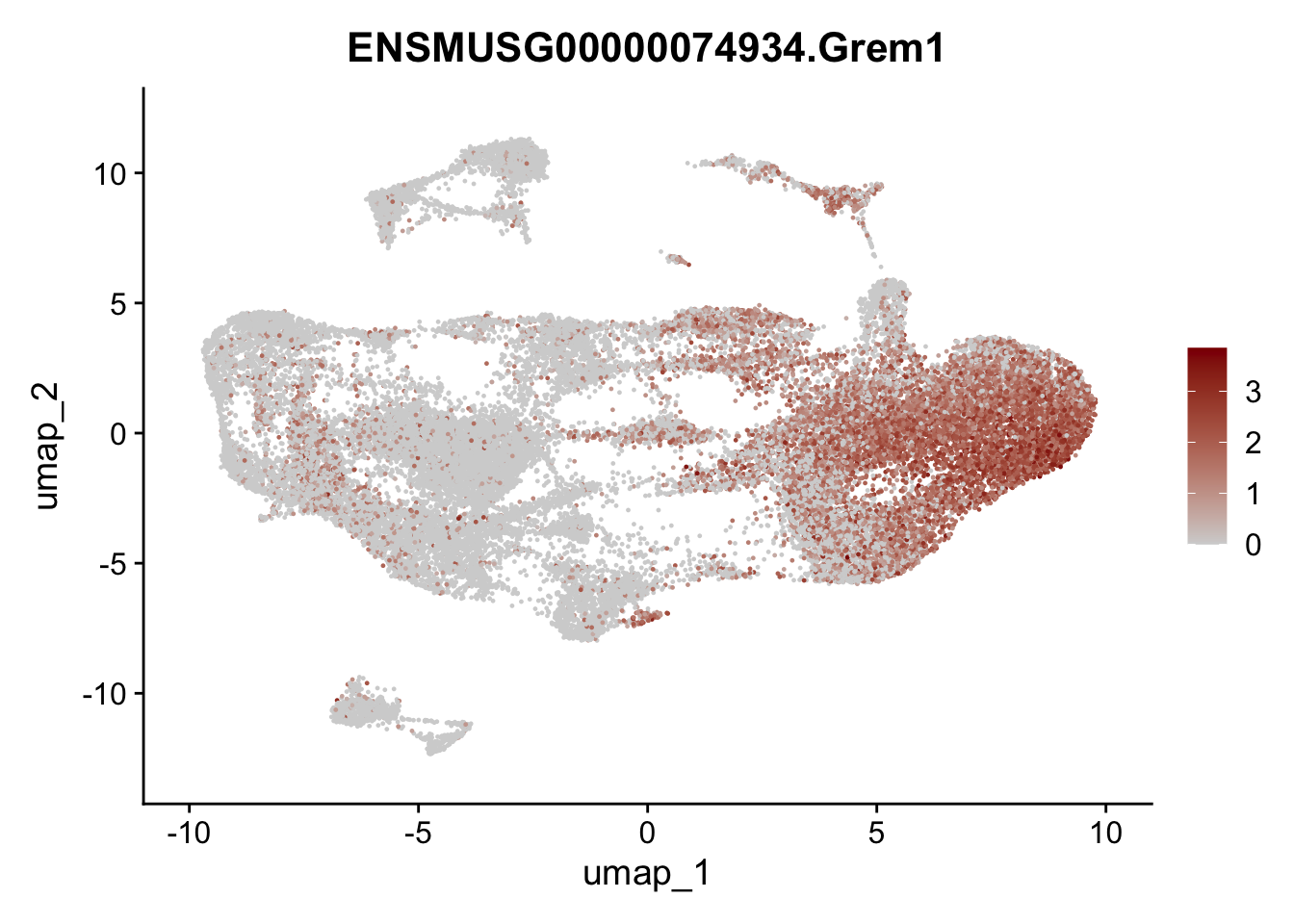
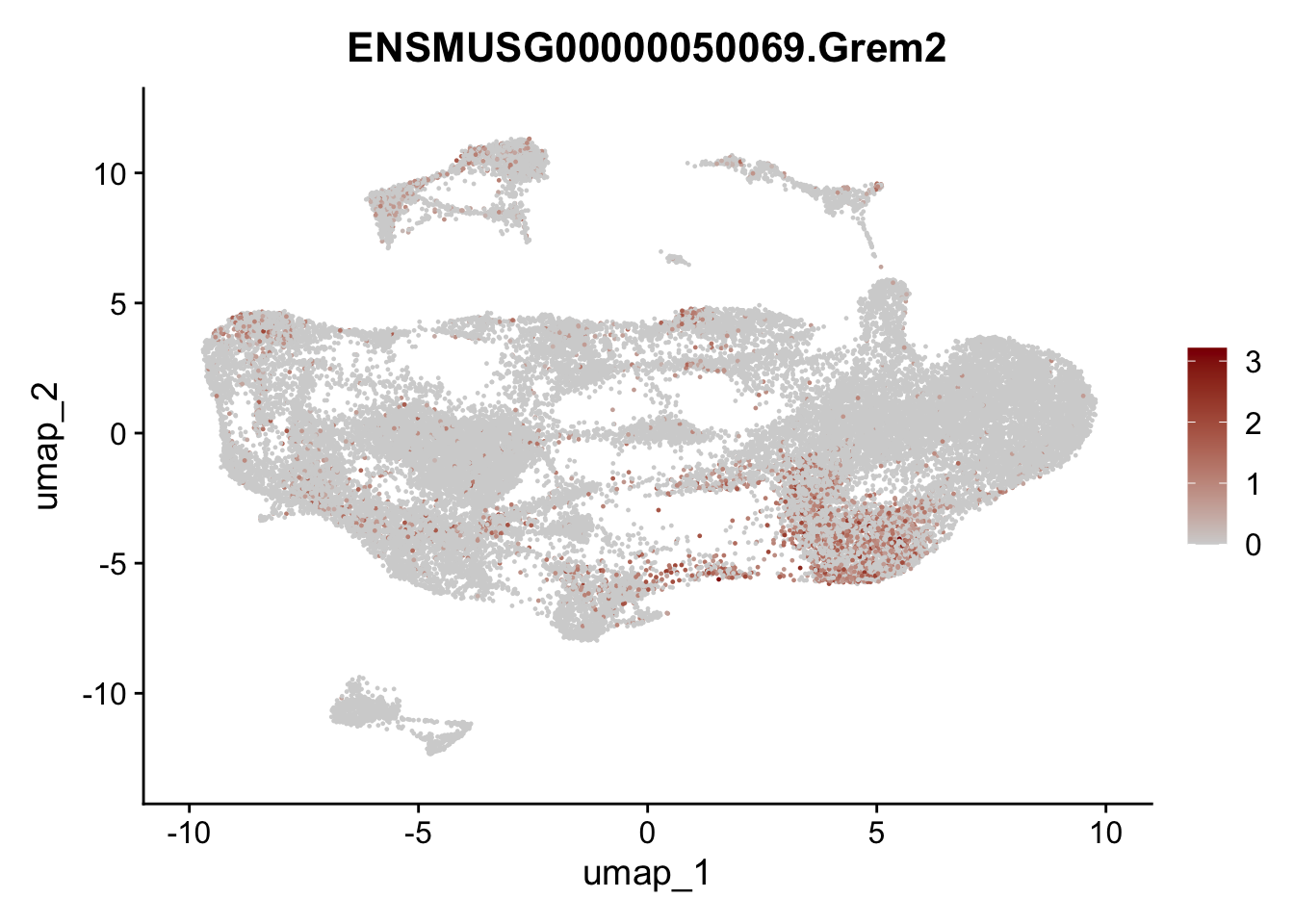
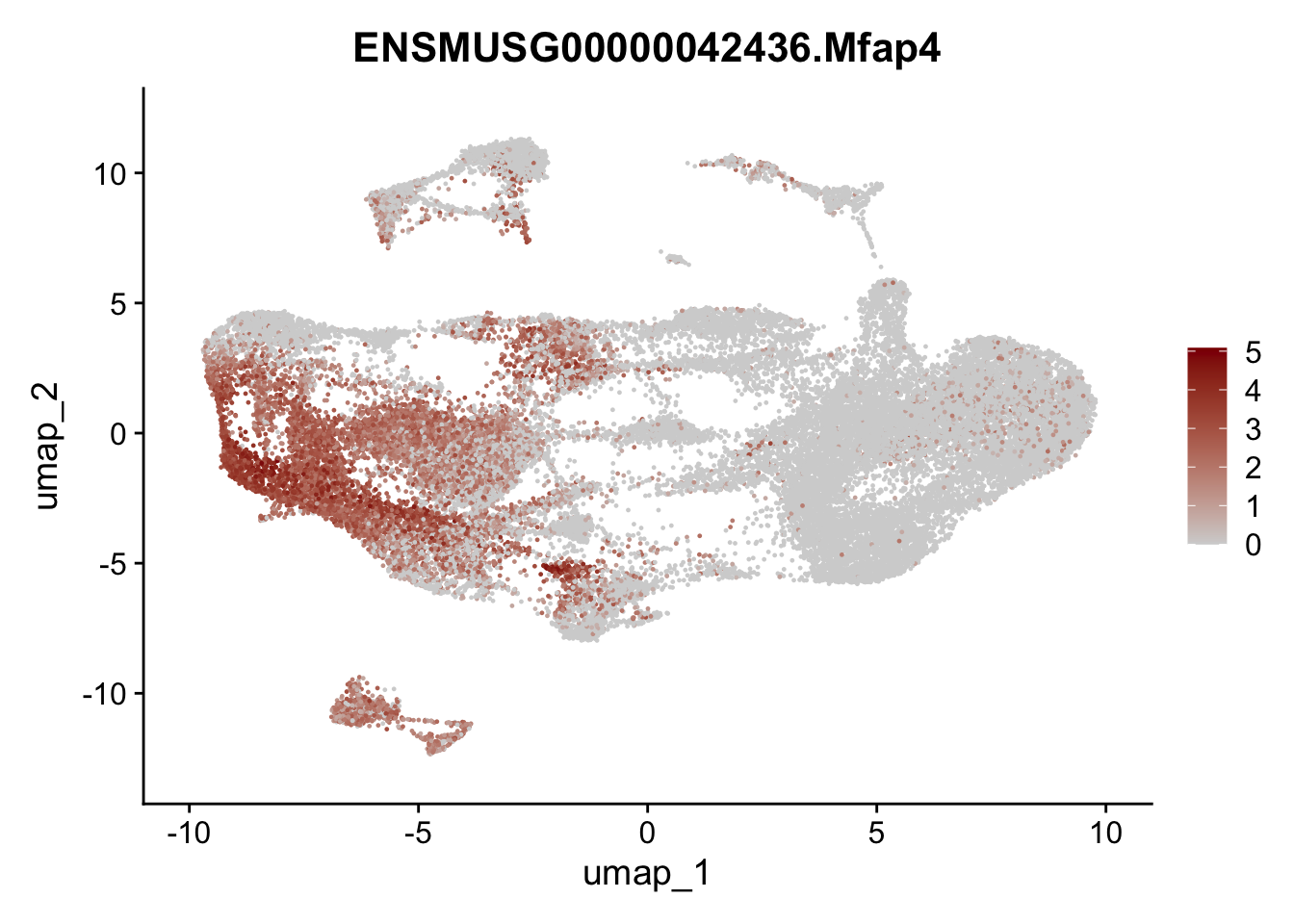
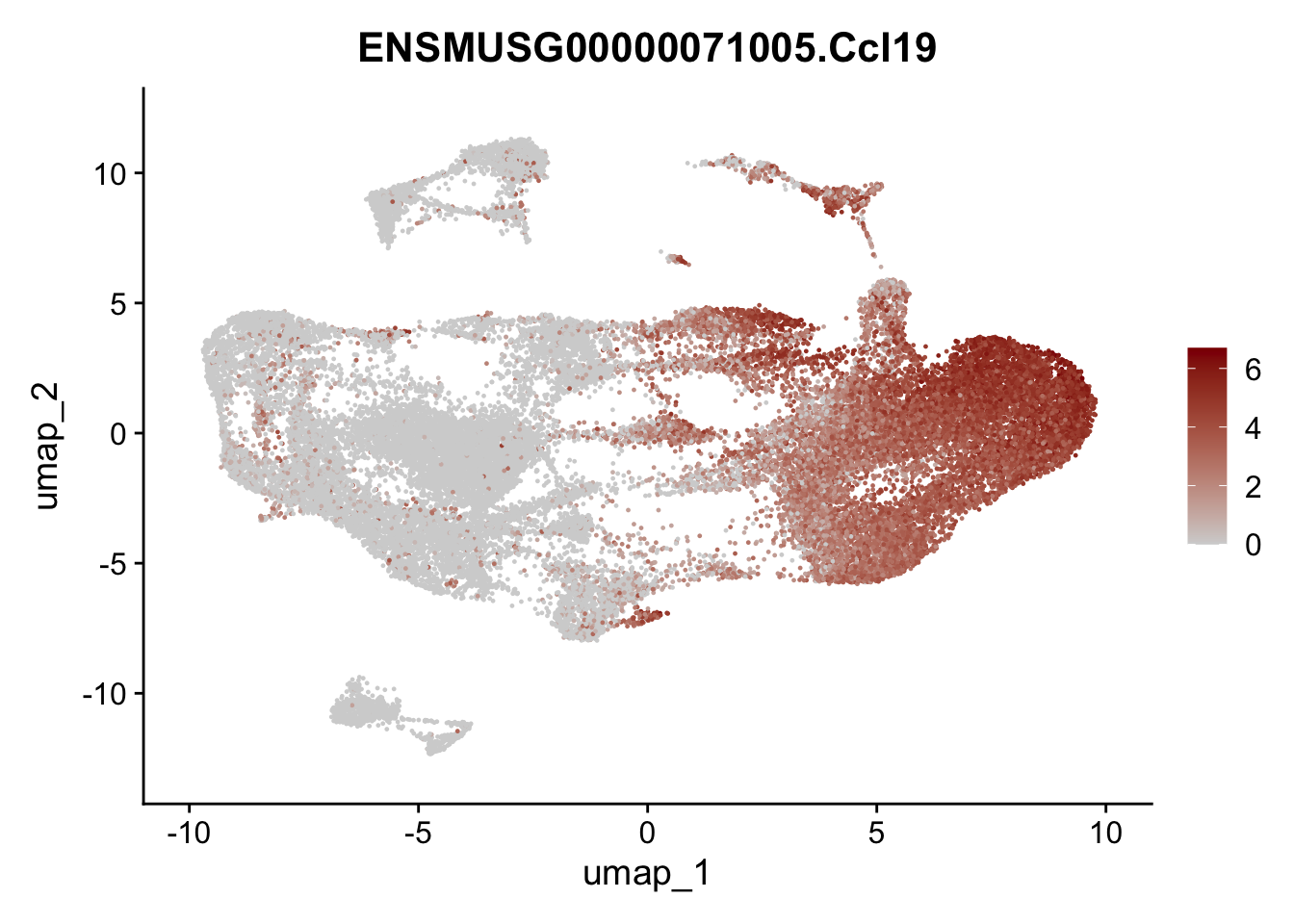
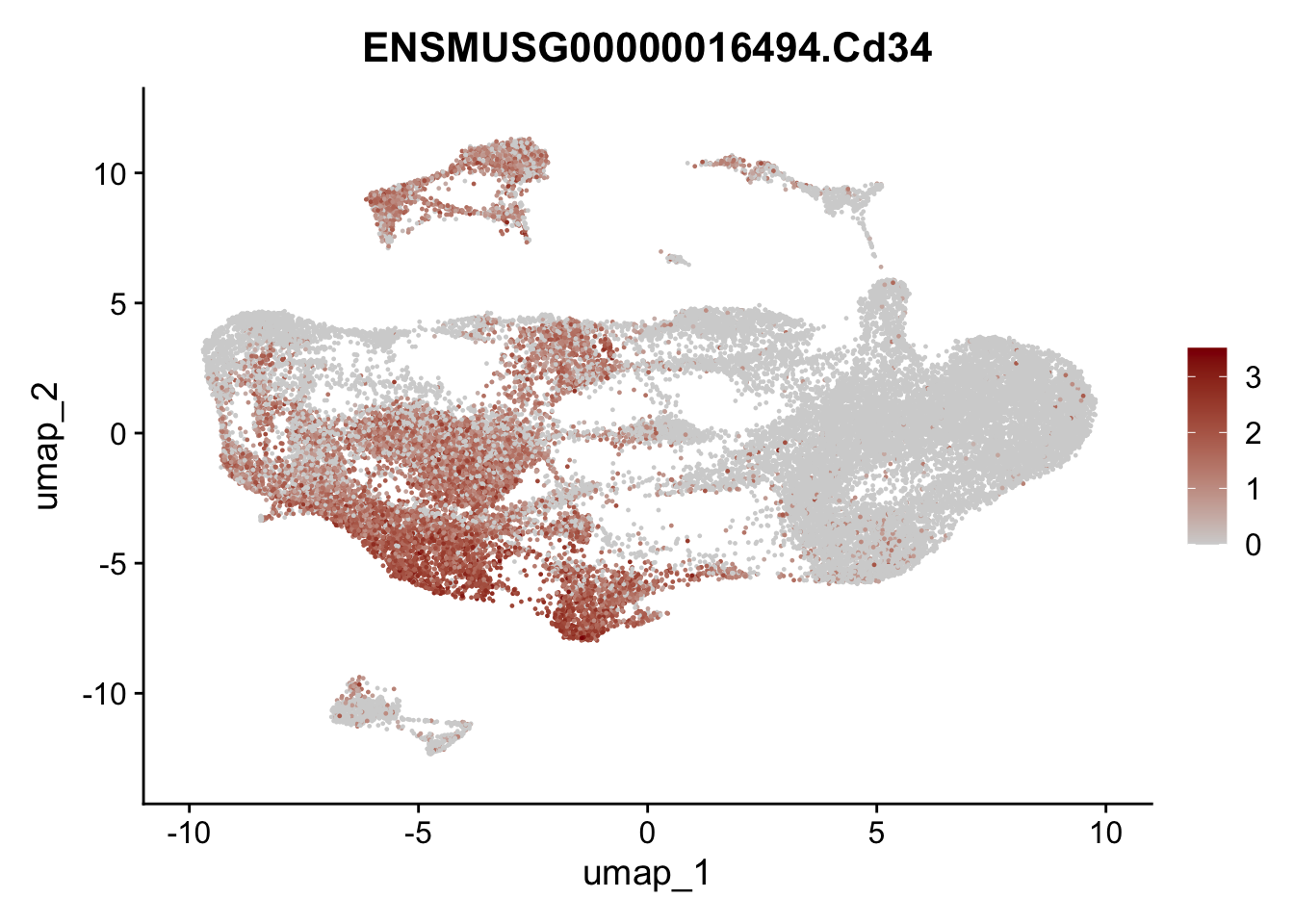
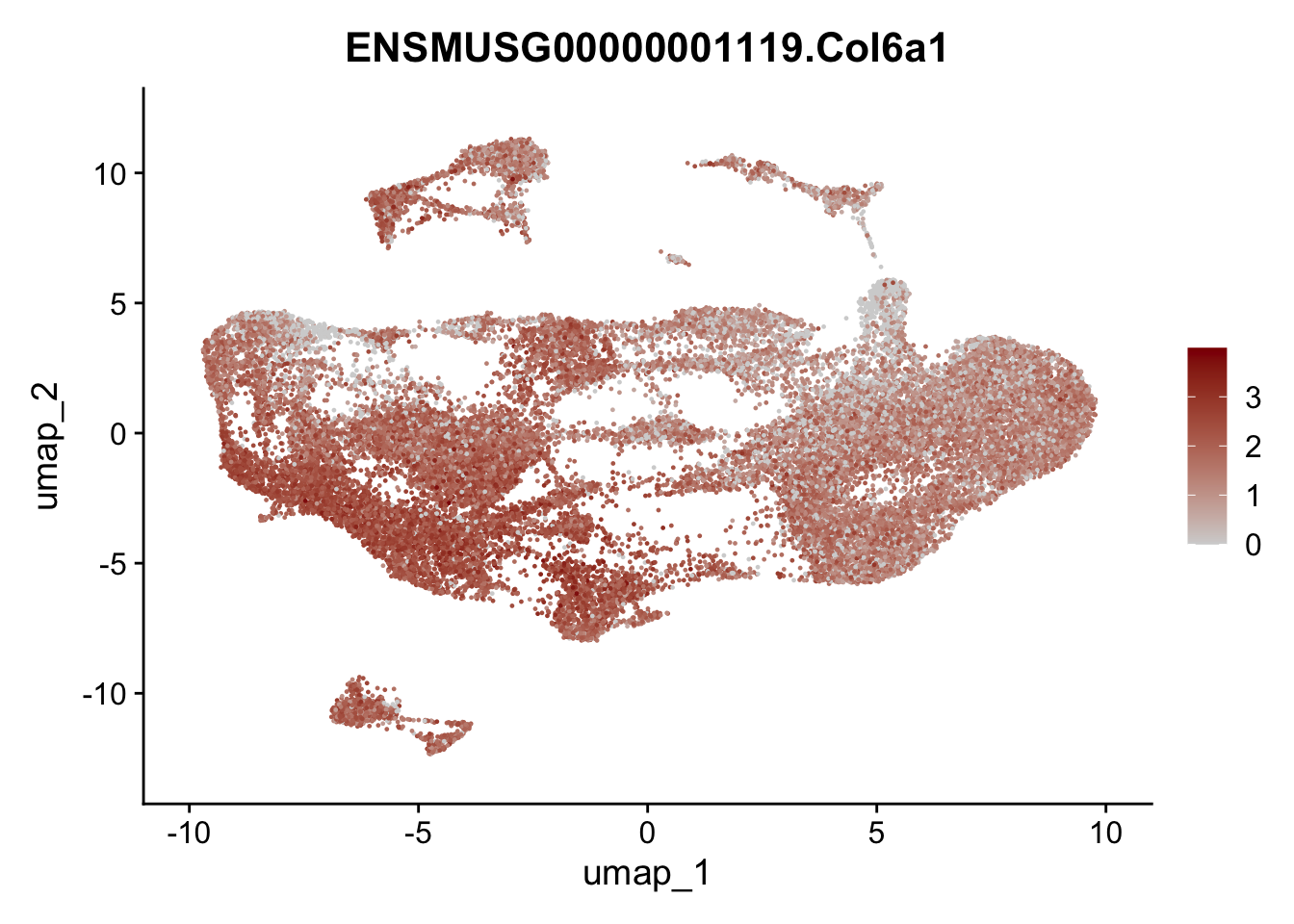
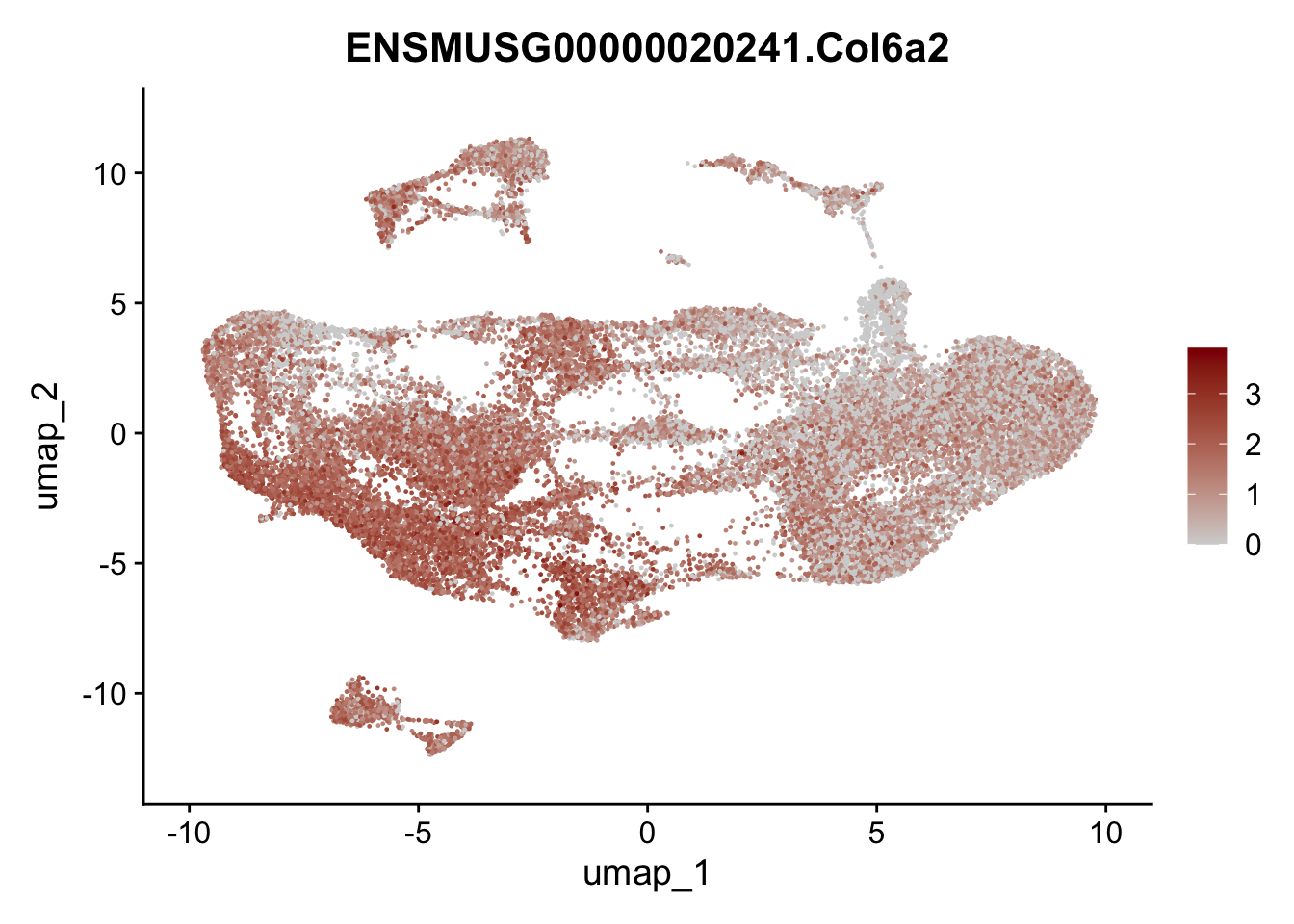
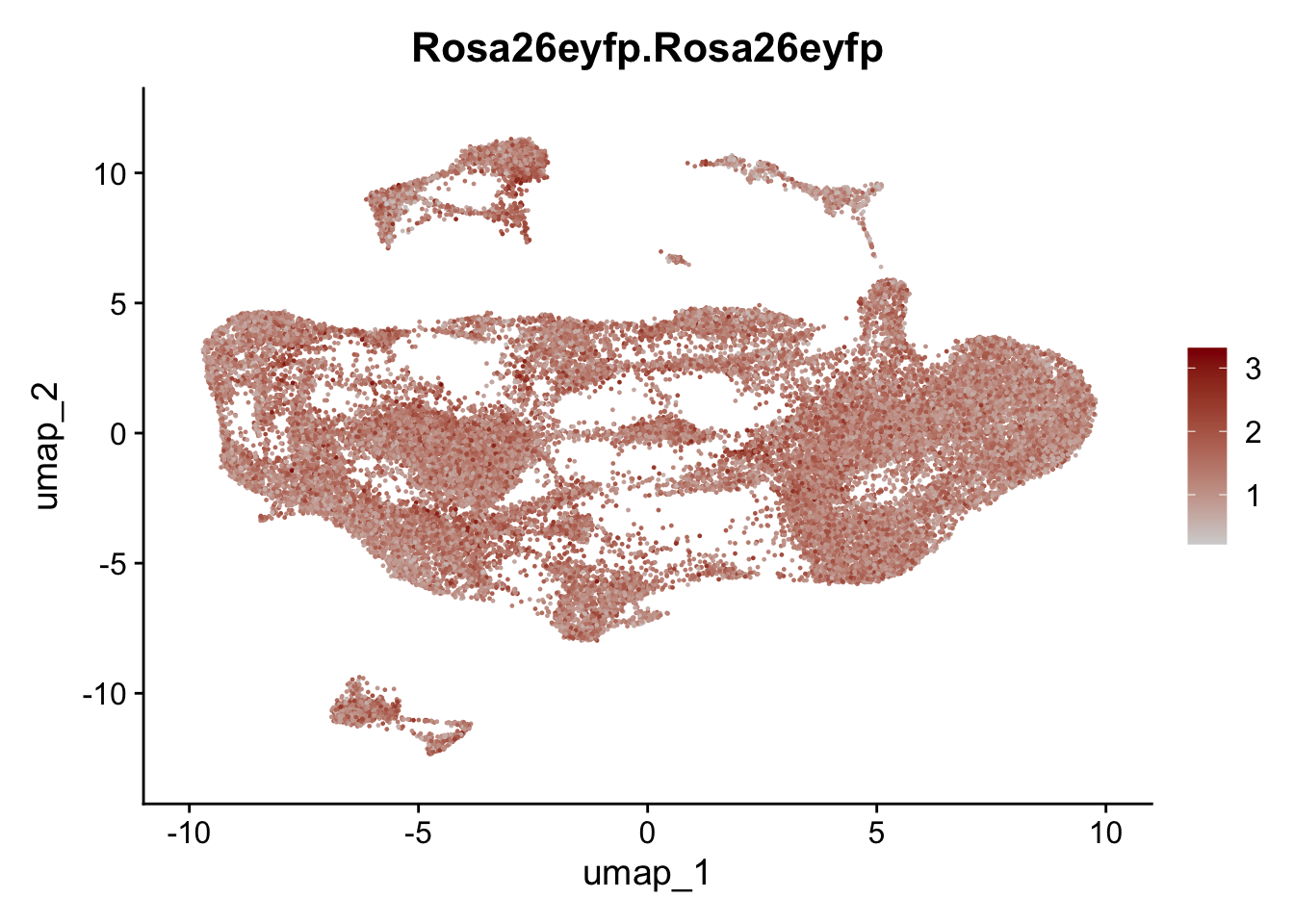
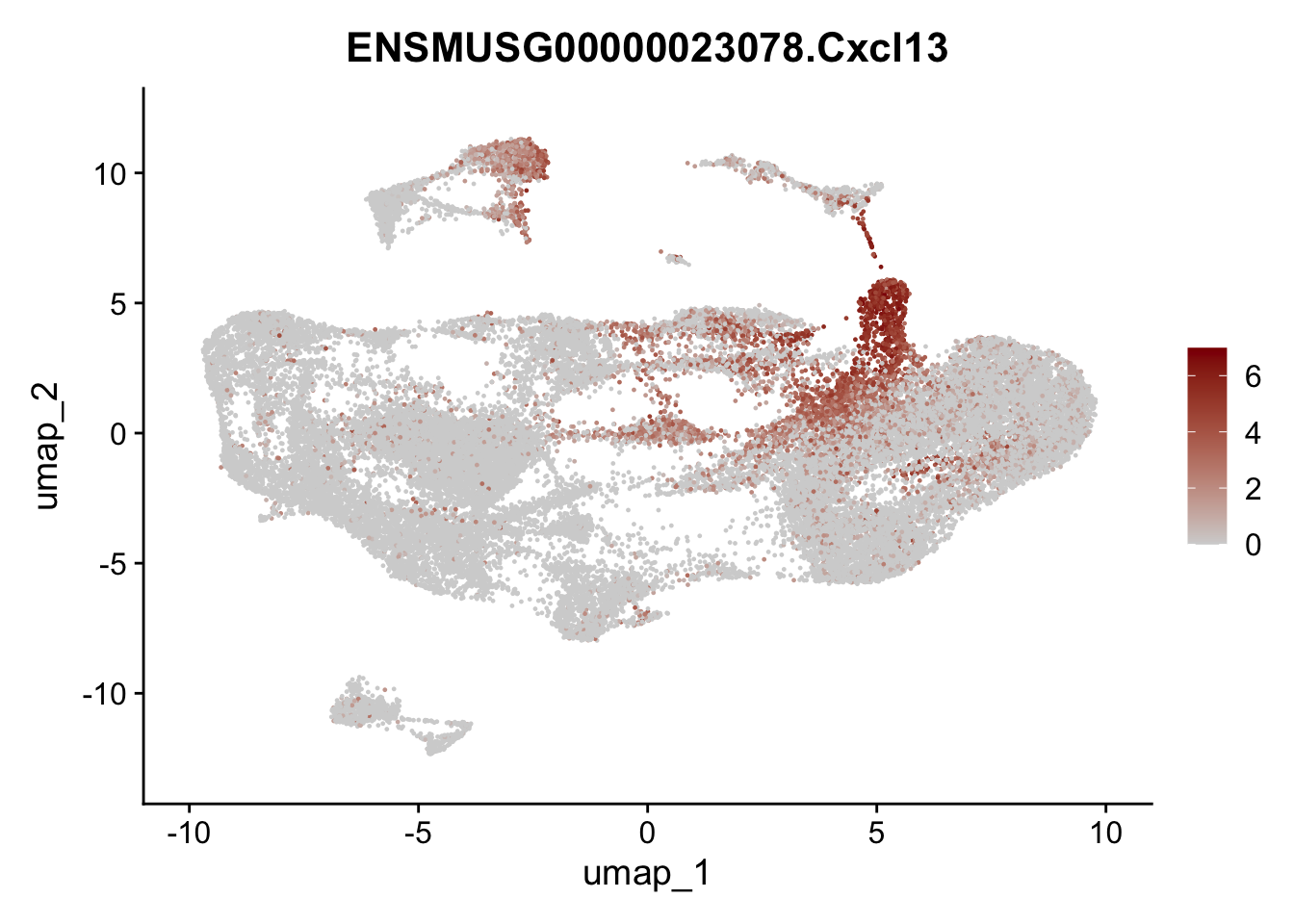
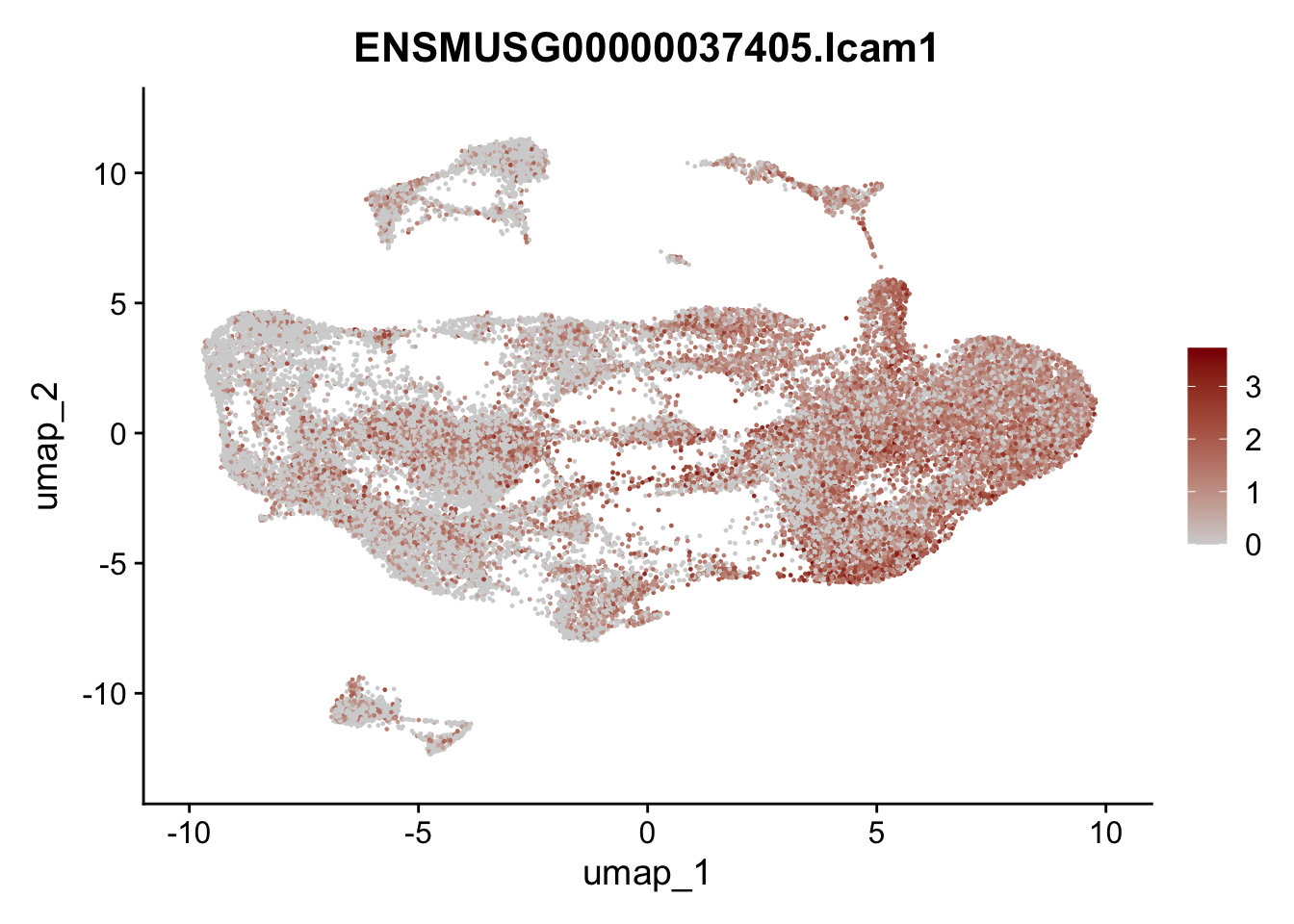
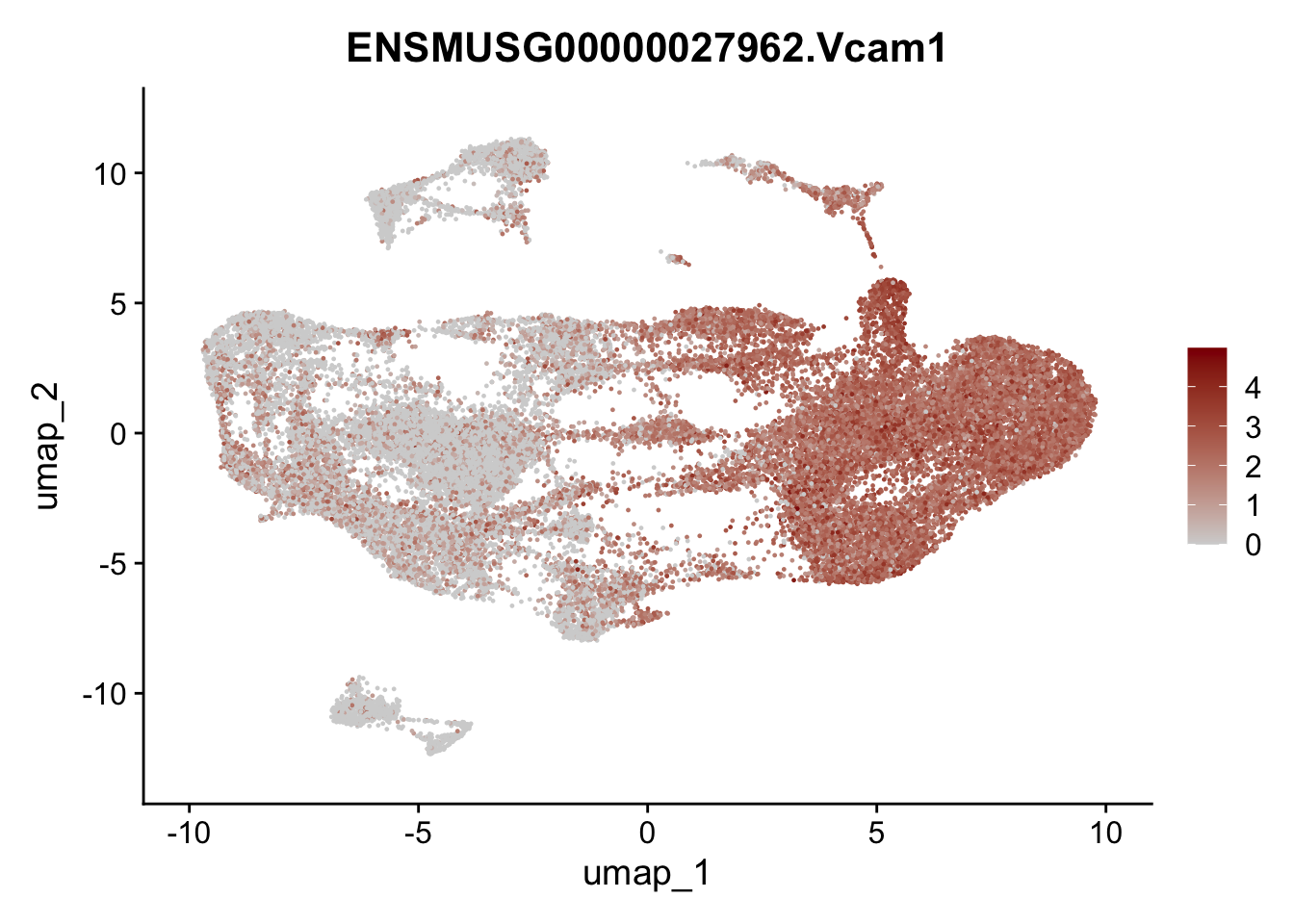
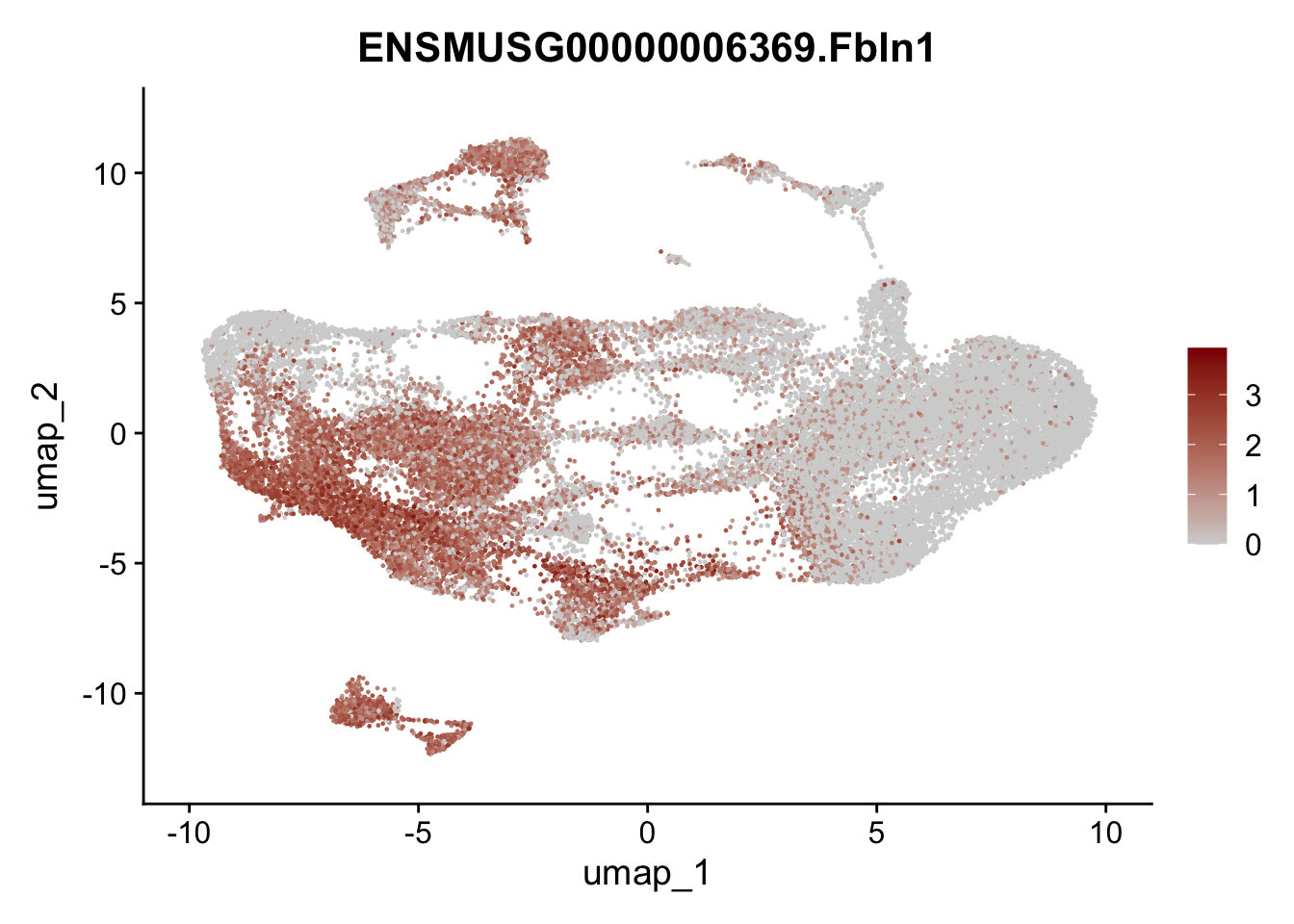
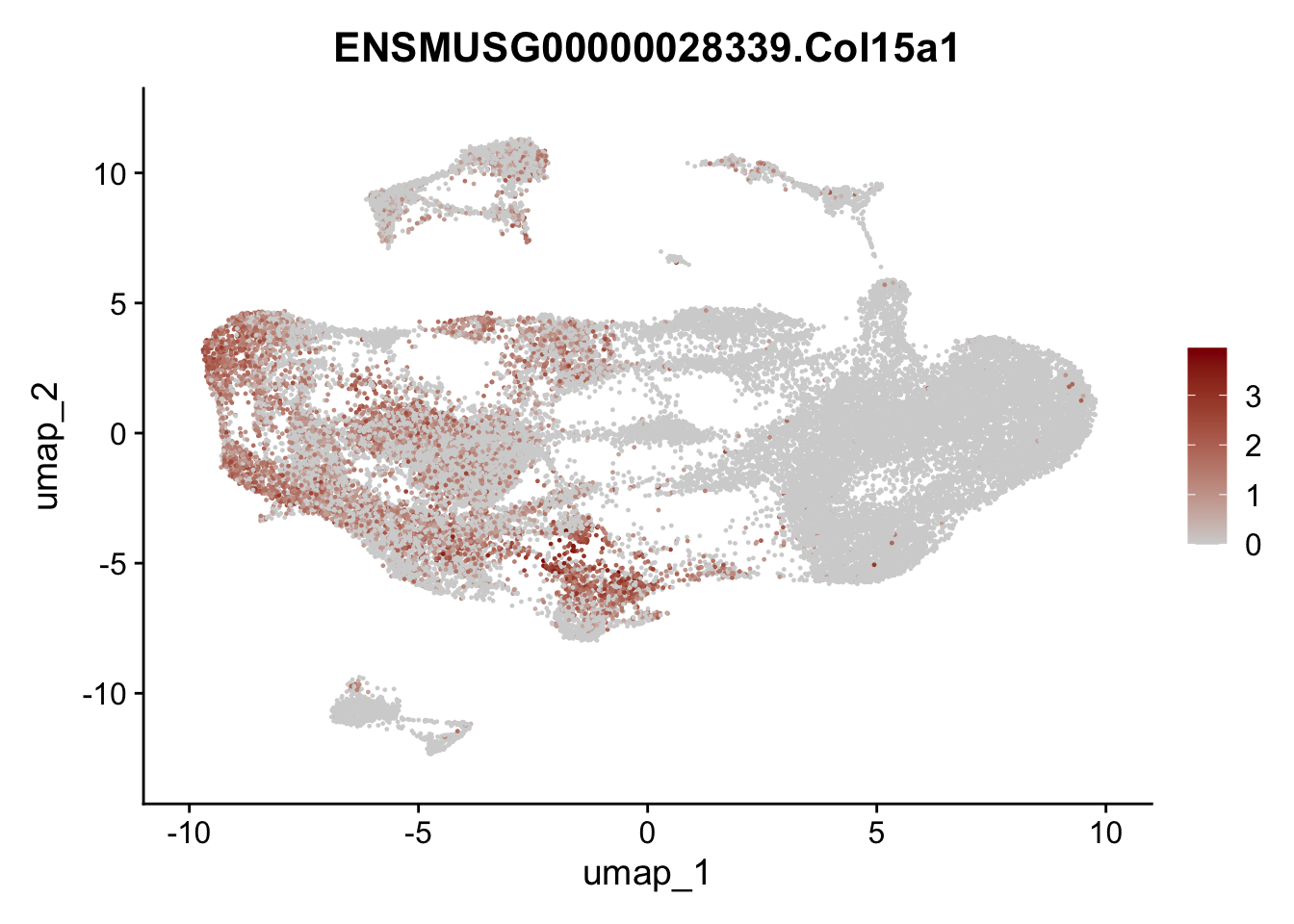
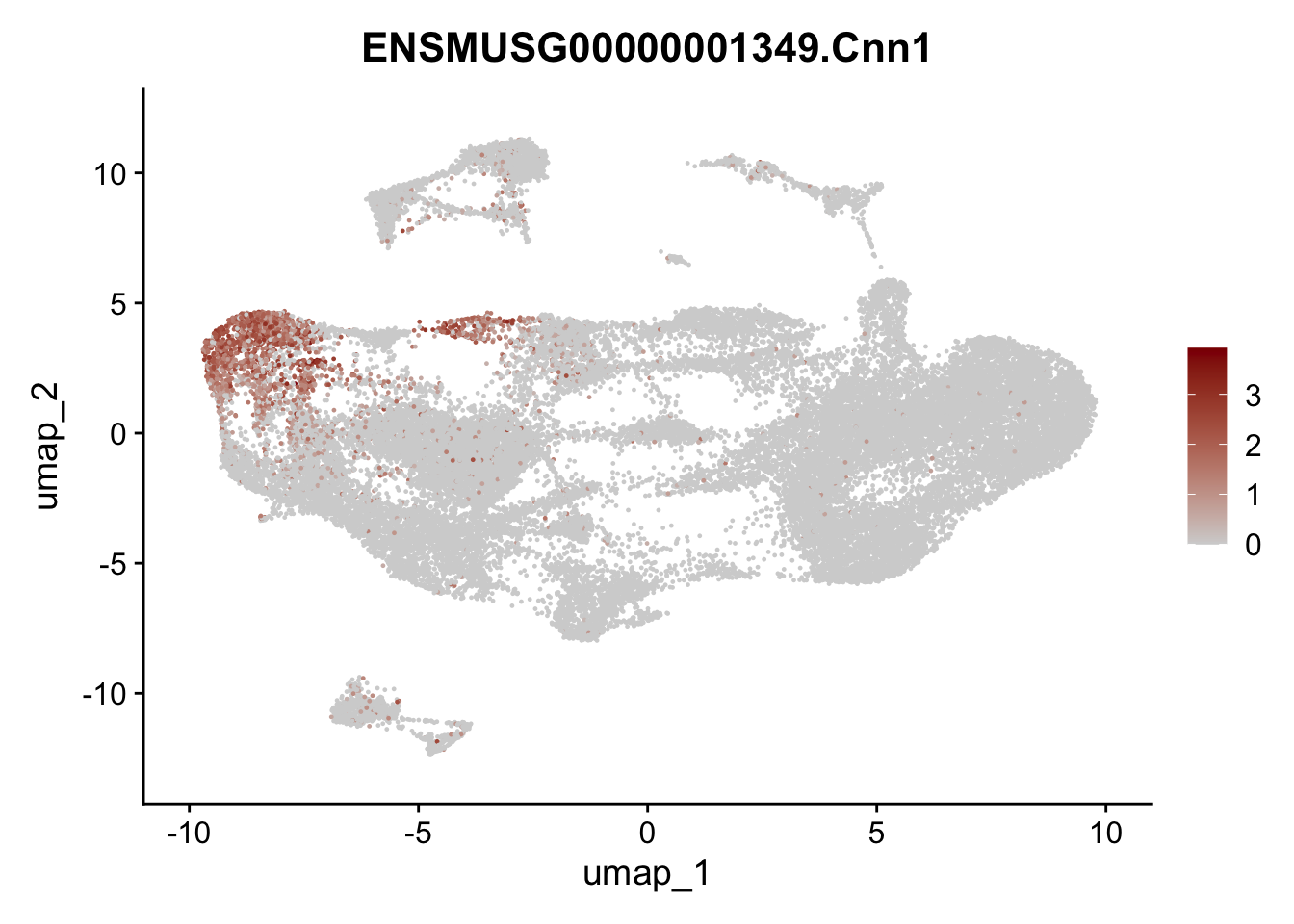
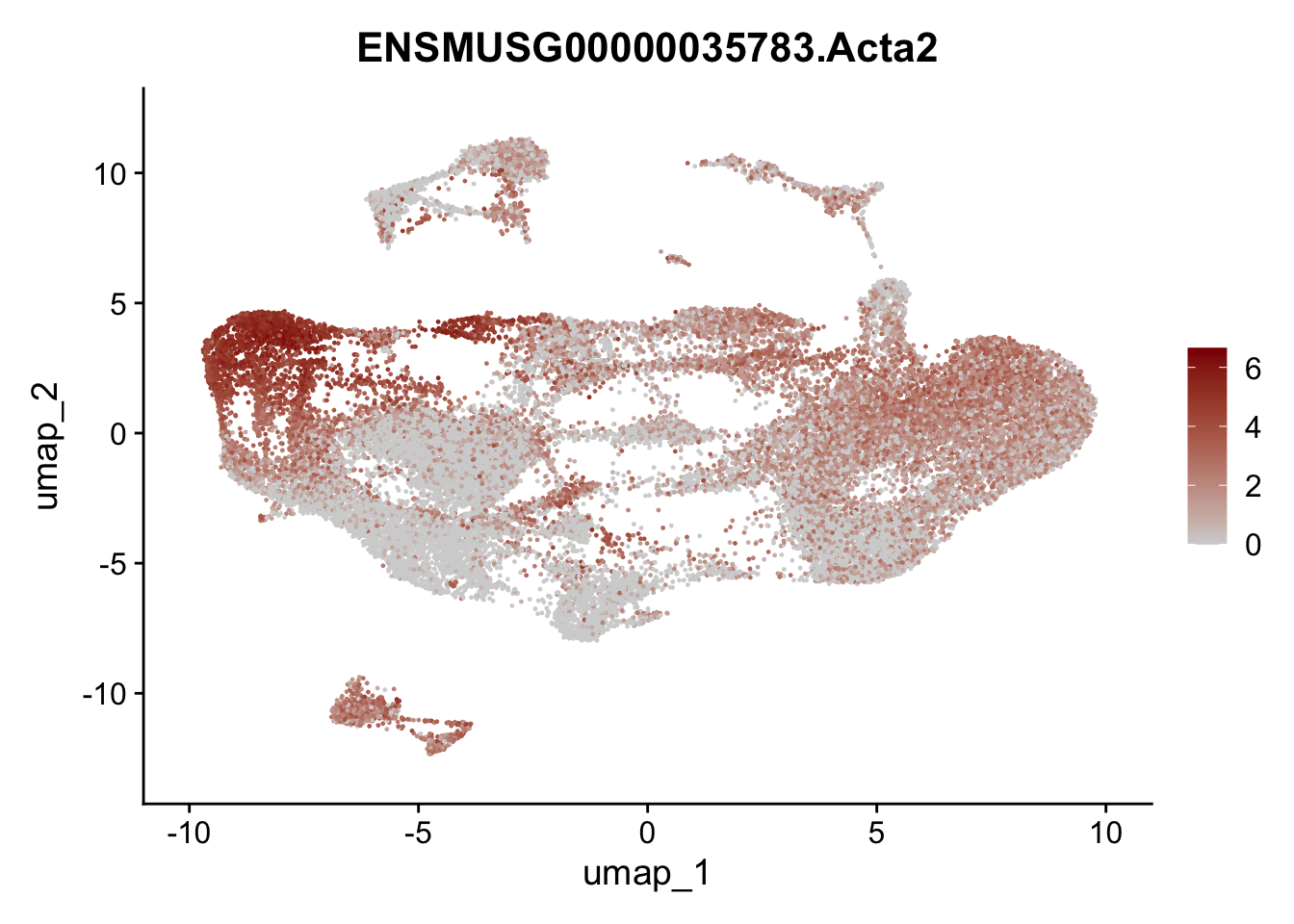
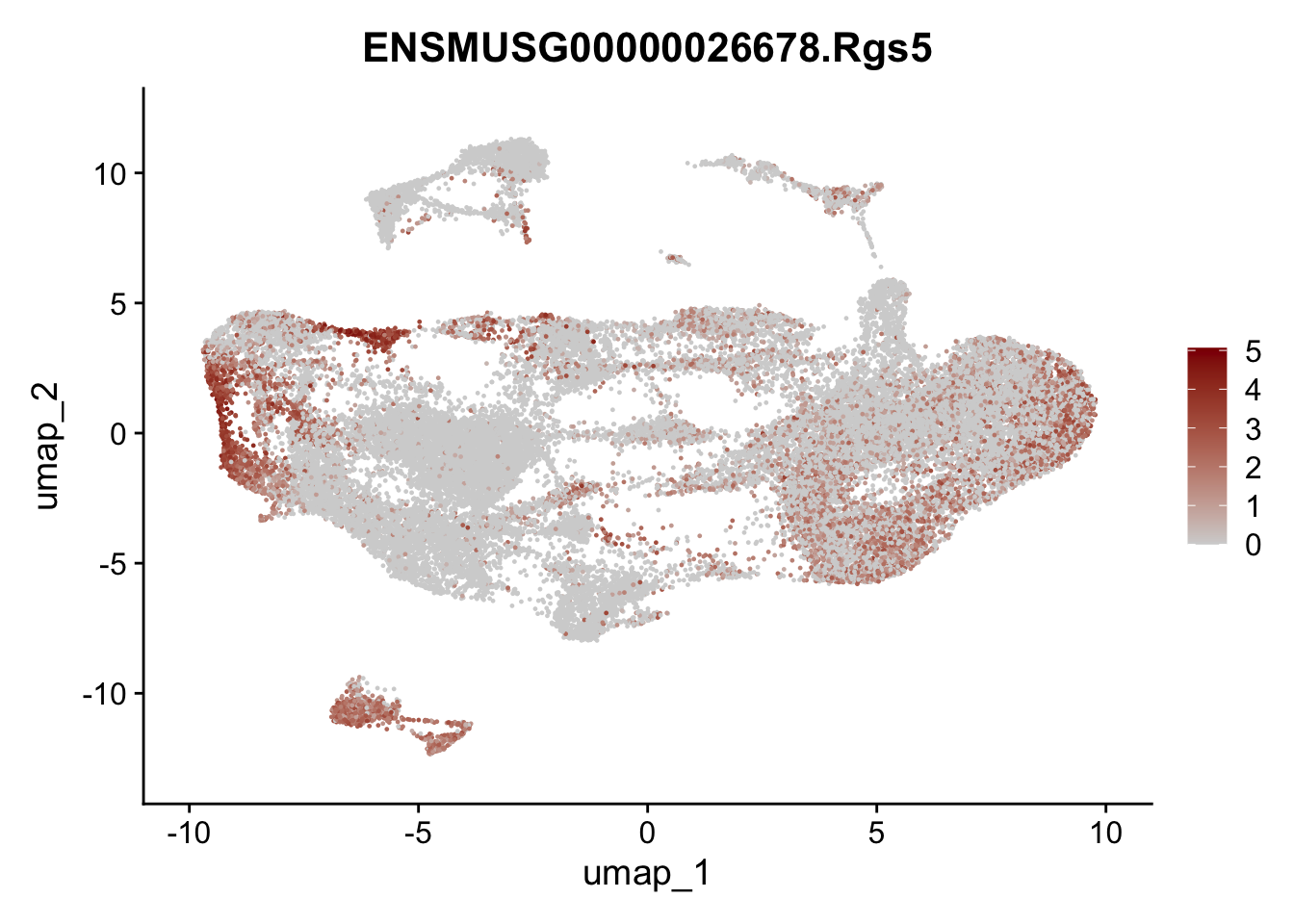
selGenesAll <- data.frame(geneID=c("Fbln1", "Col15a1", "Cnn1", "Acta2", "Rgs5",
"Cox4i2", "Pi16", "Cd34", "Emp1", "Ogn",
"Fhl2")) %>%
left_join(., genes, by = "geneID")
pList <- sapply(selGenesAll$gene, function(x){
p <- FeaturePlot(seuratmLN, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = F)+
theme(legend.position="right")
plot(p)
})
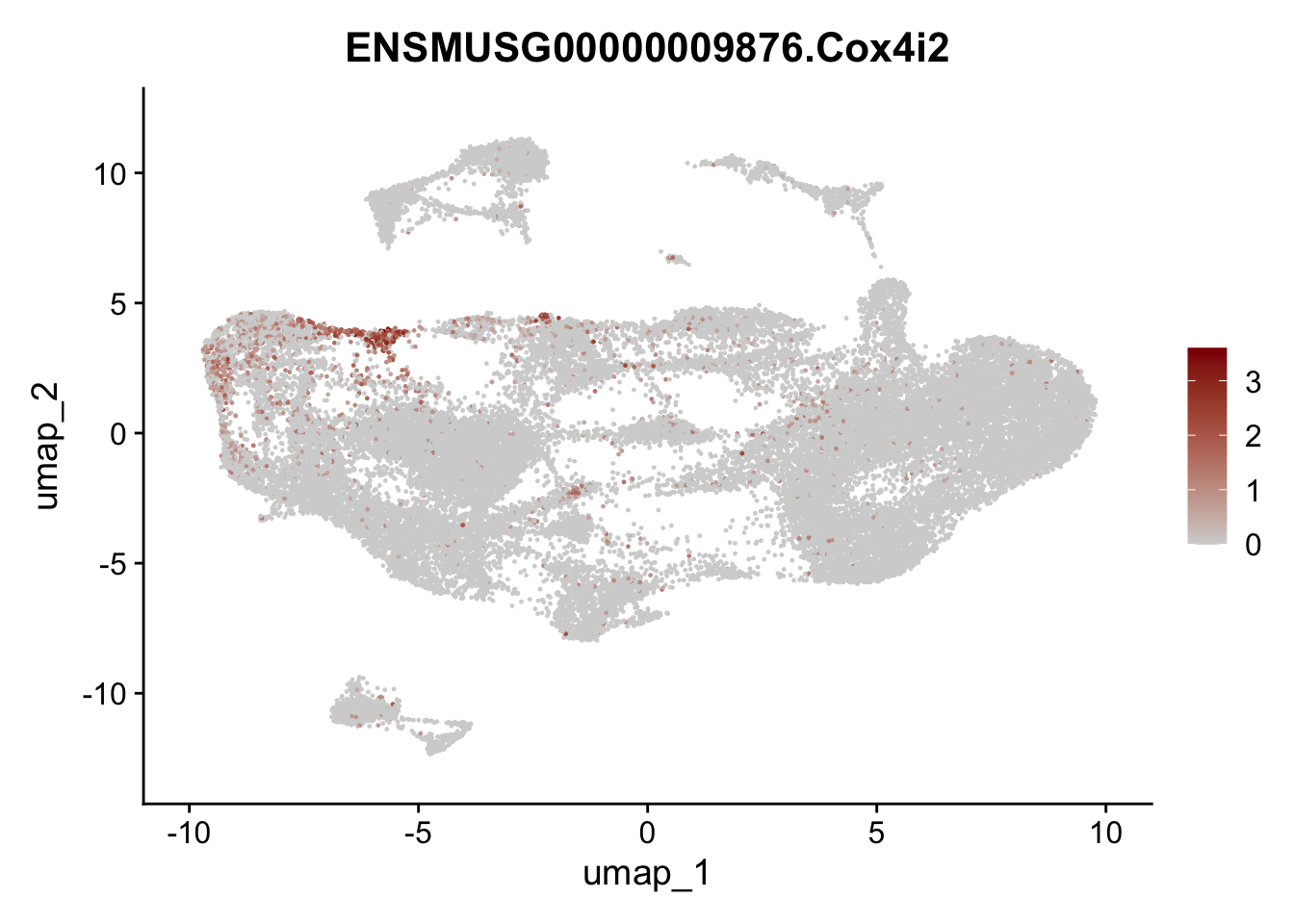
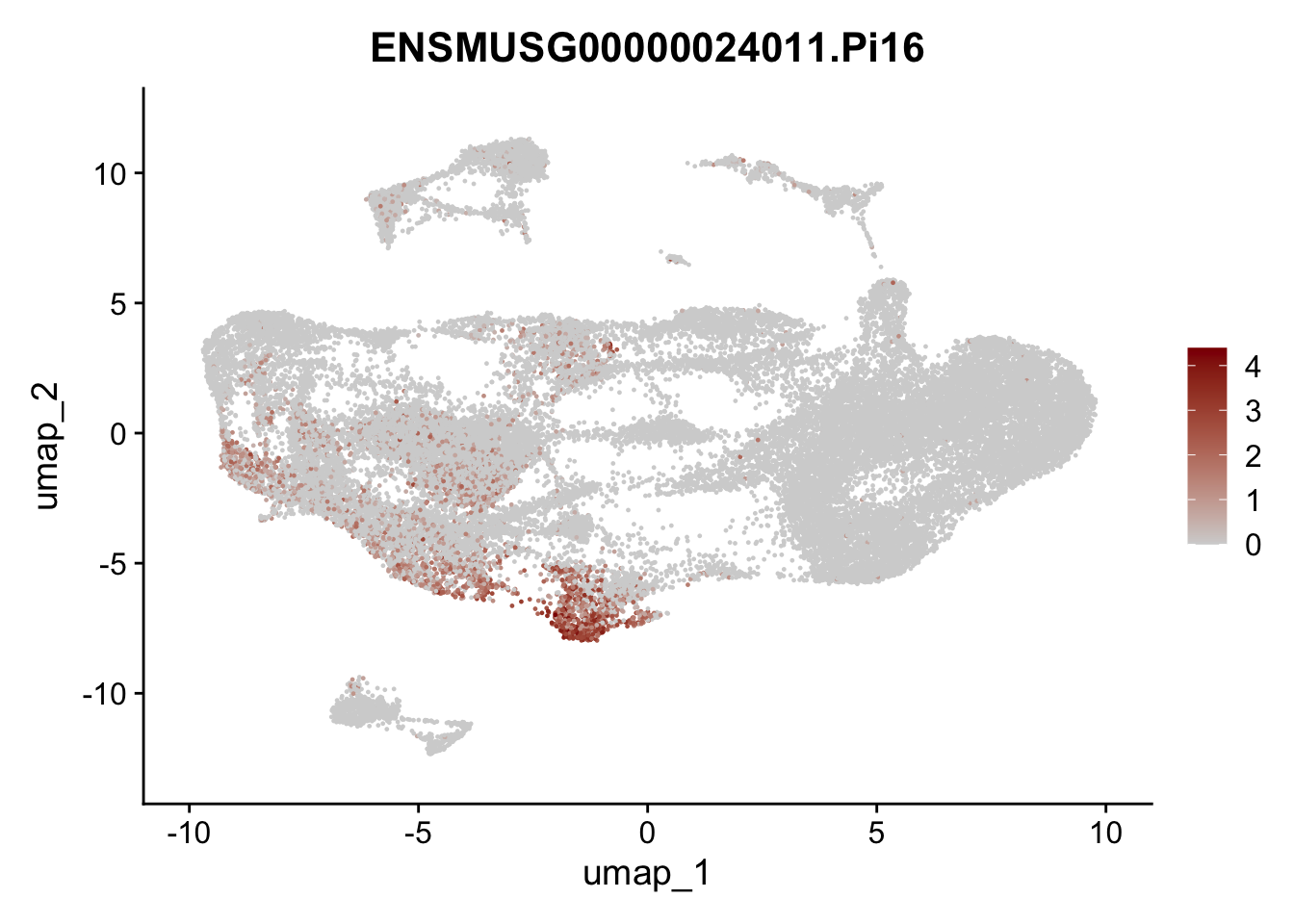
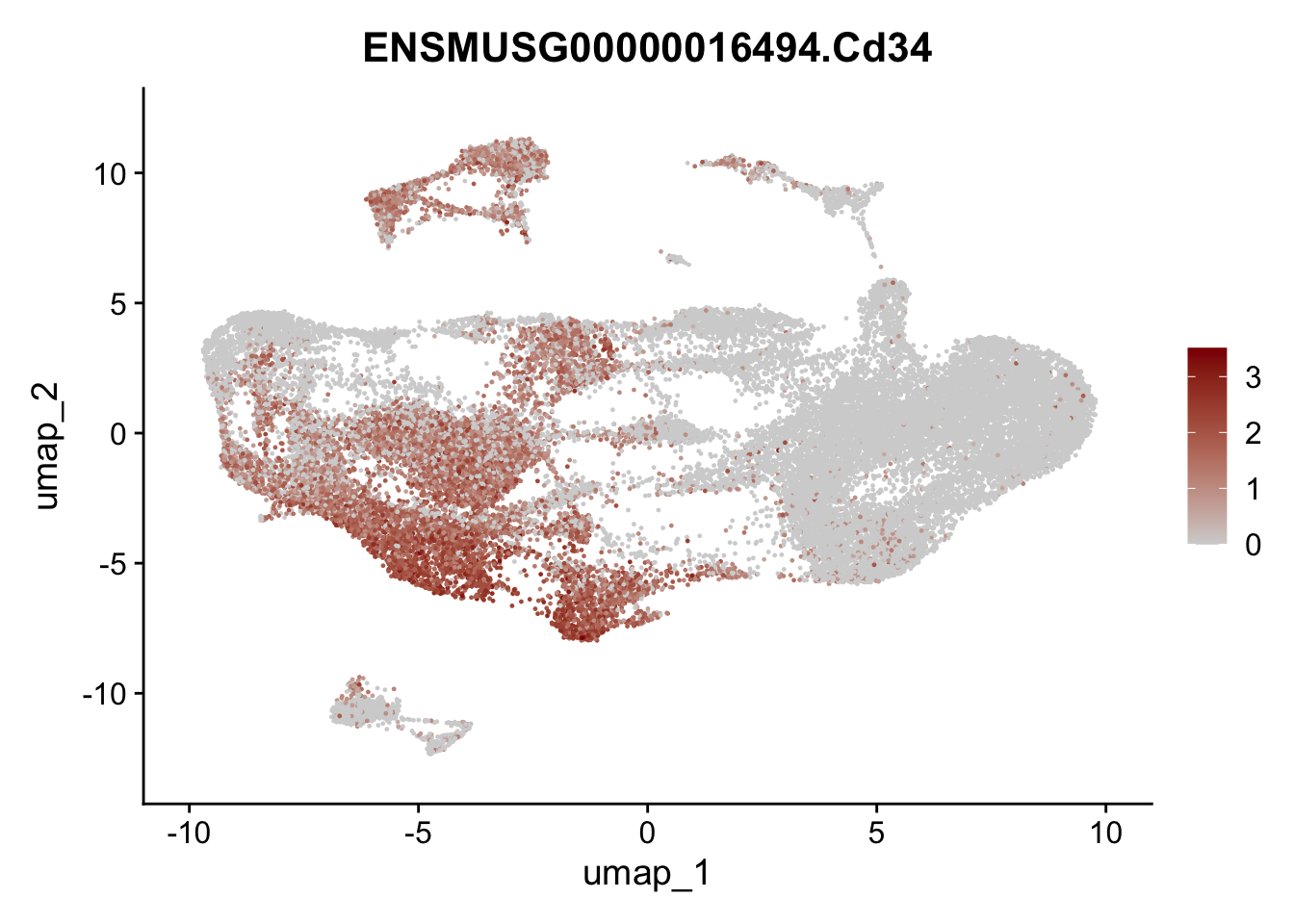
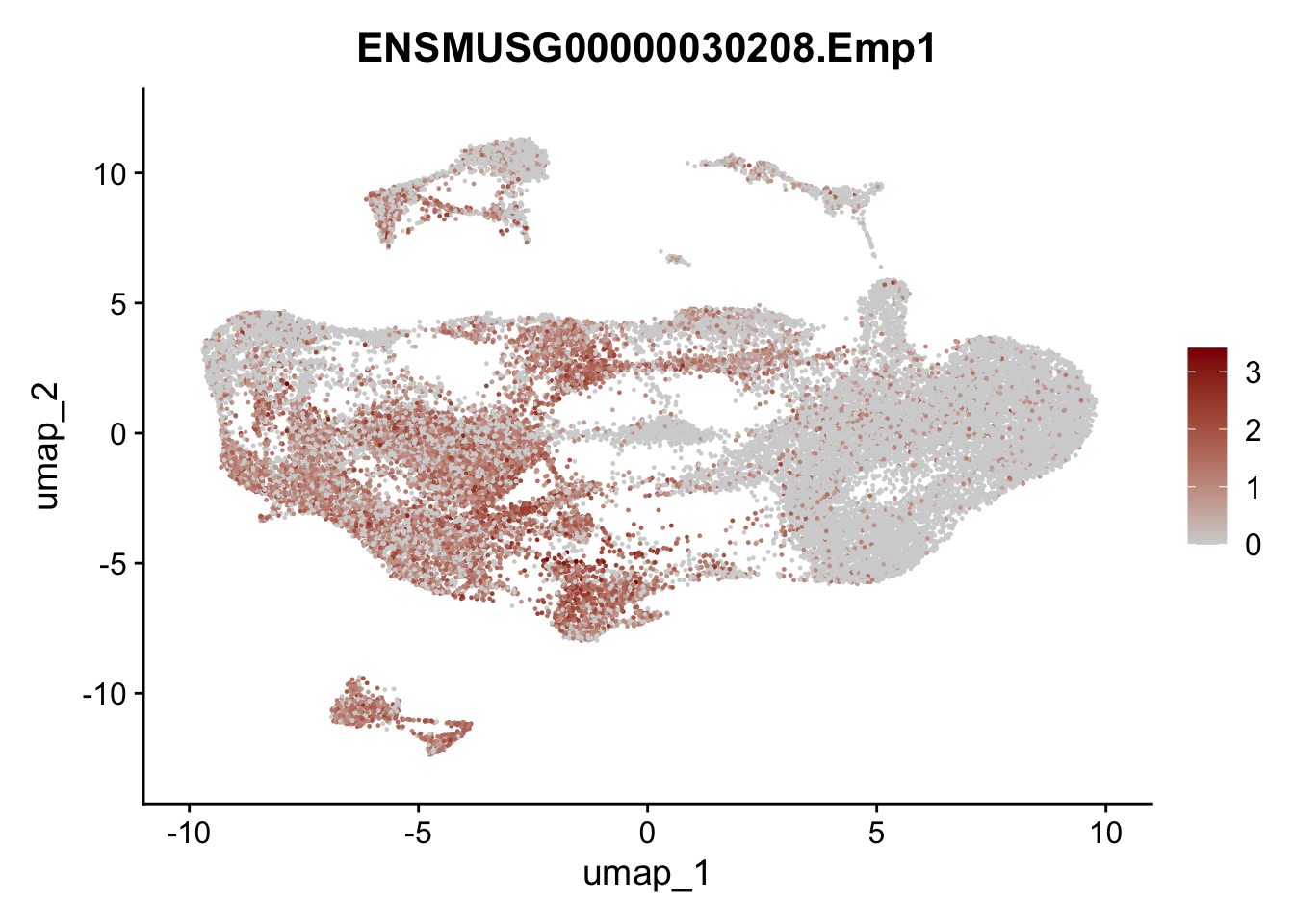
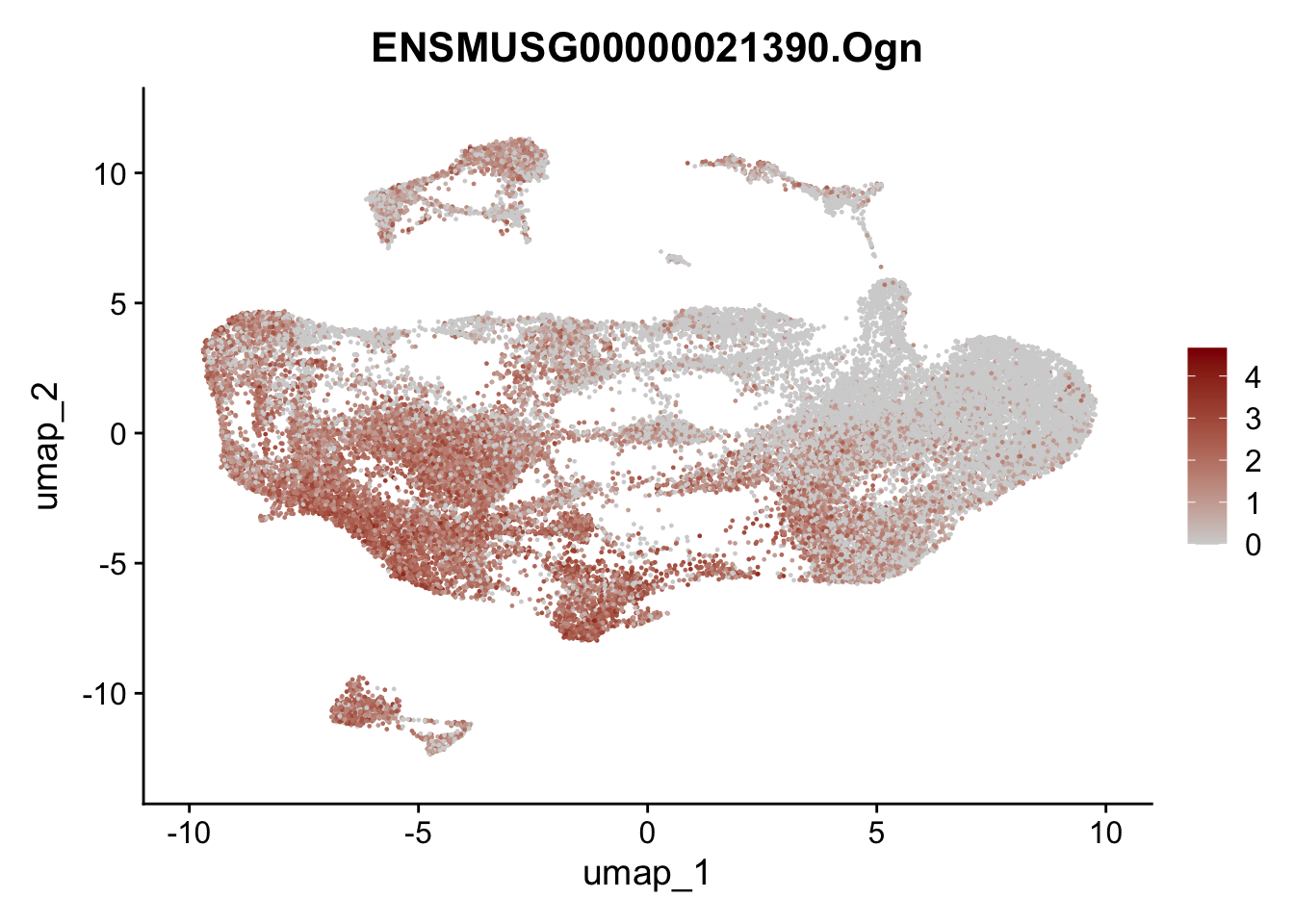
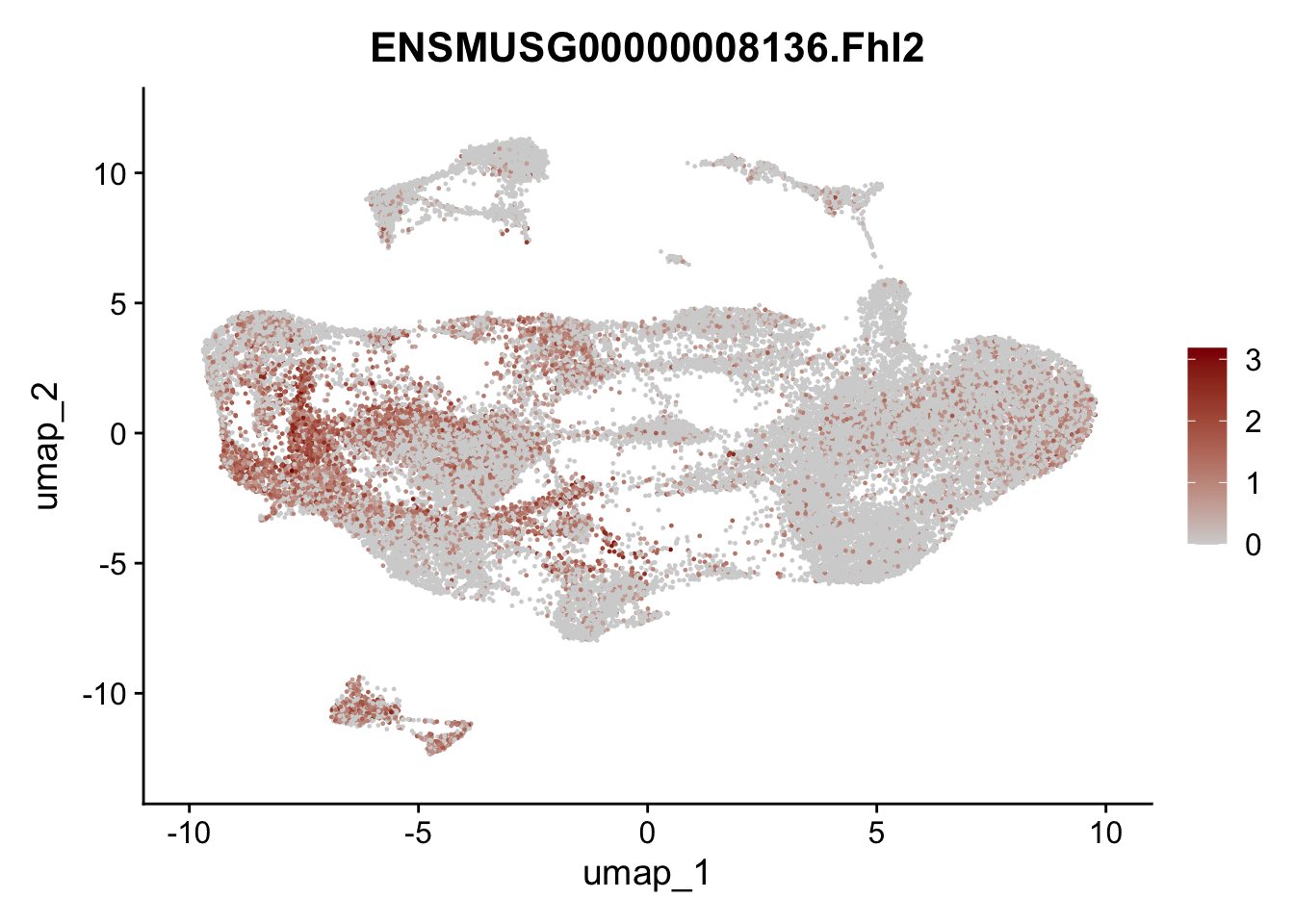
selGenesfil <- c("ENSMUSG00000026395.Ptprc", "ENSMUSG00000031004.Mki67", "ENSMUSG00000063011.Msln", "ENSMUSG00000045680.Tcf21")
pList <- sapply(selGenesfil, function(x){
p <- FeaturePlot(seuratmLN, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = T)+
theme(legend.position="right")
plot(p)
})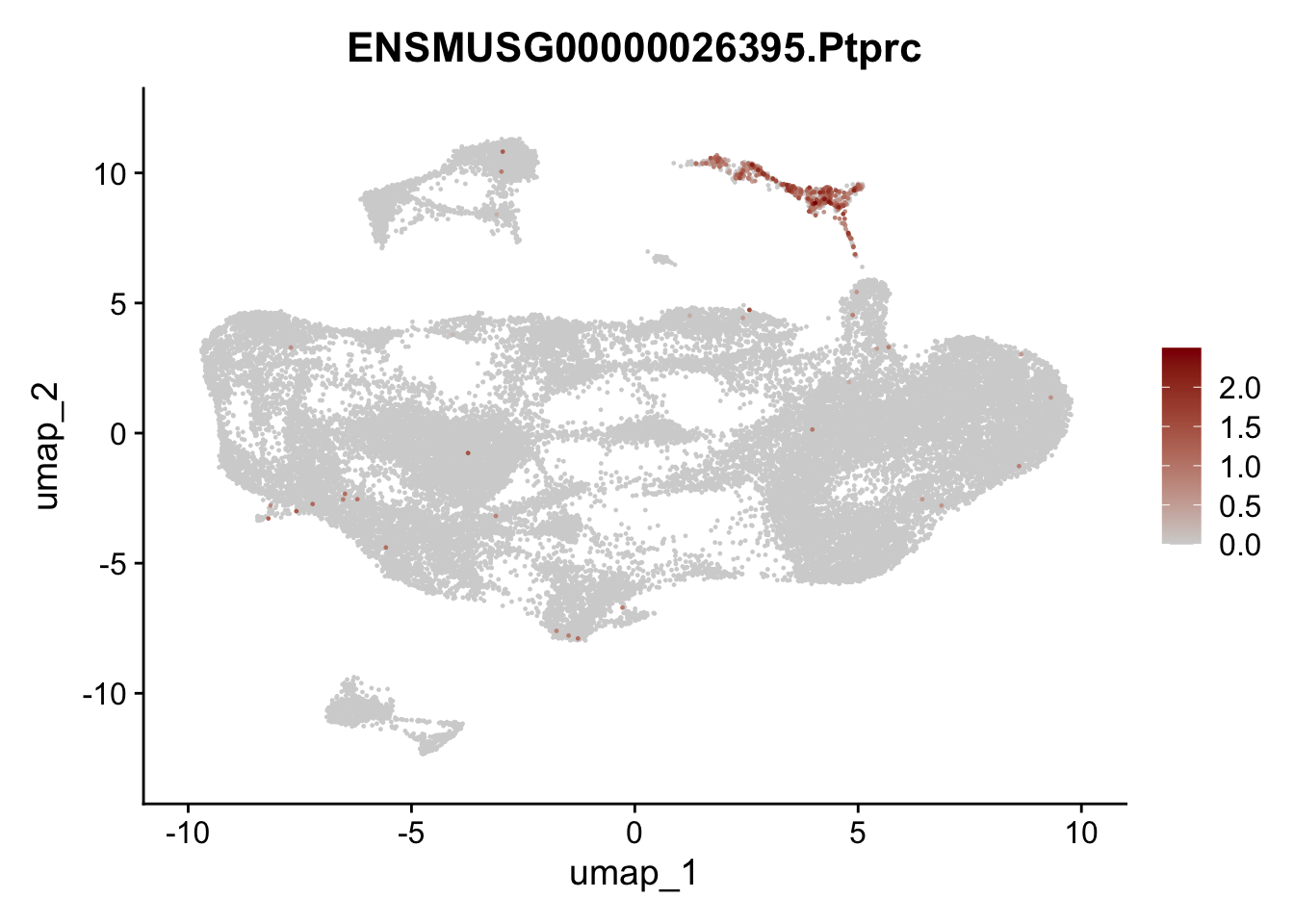
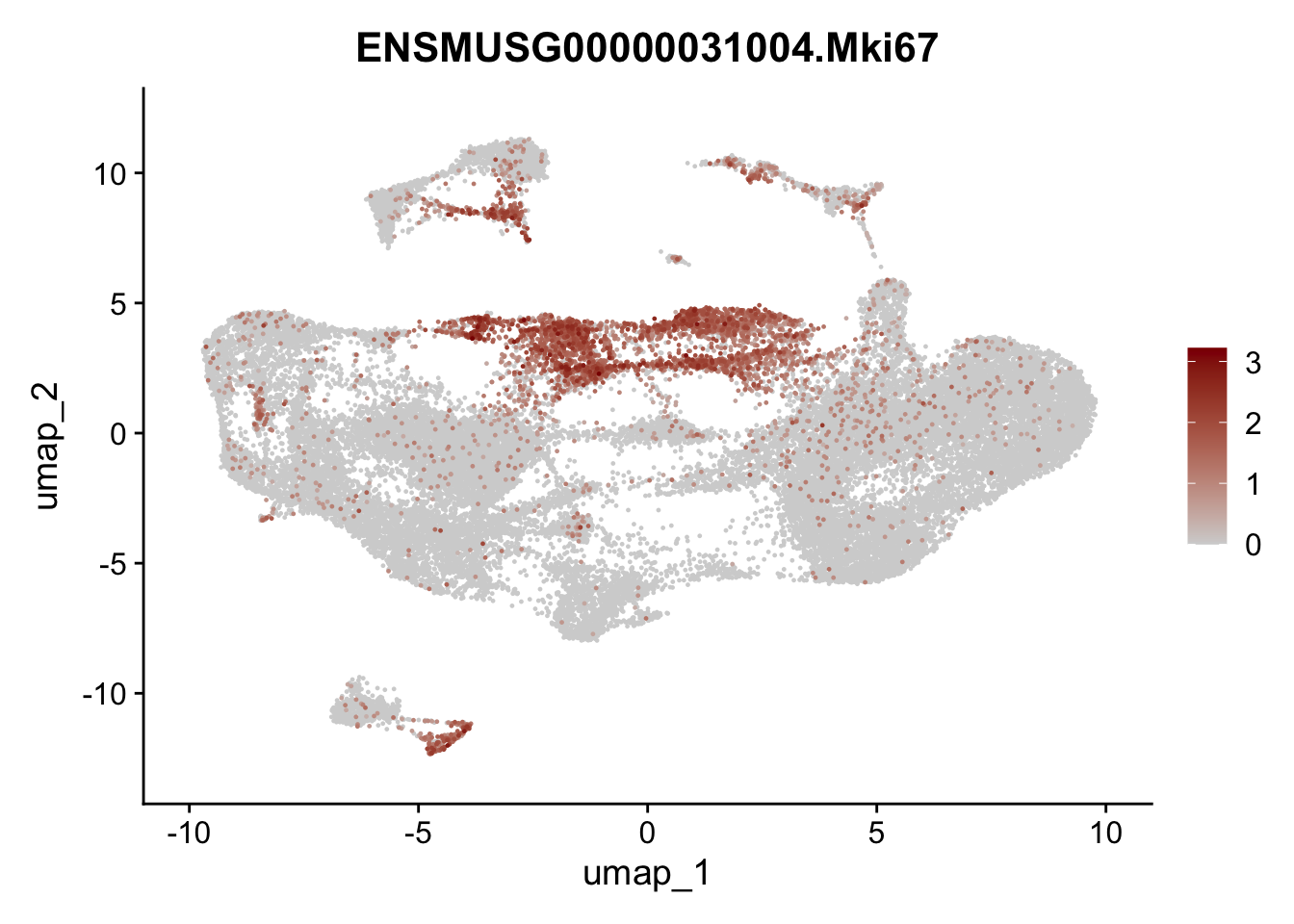
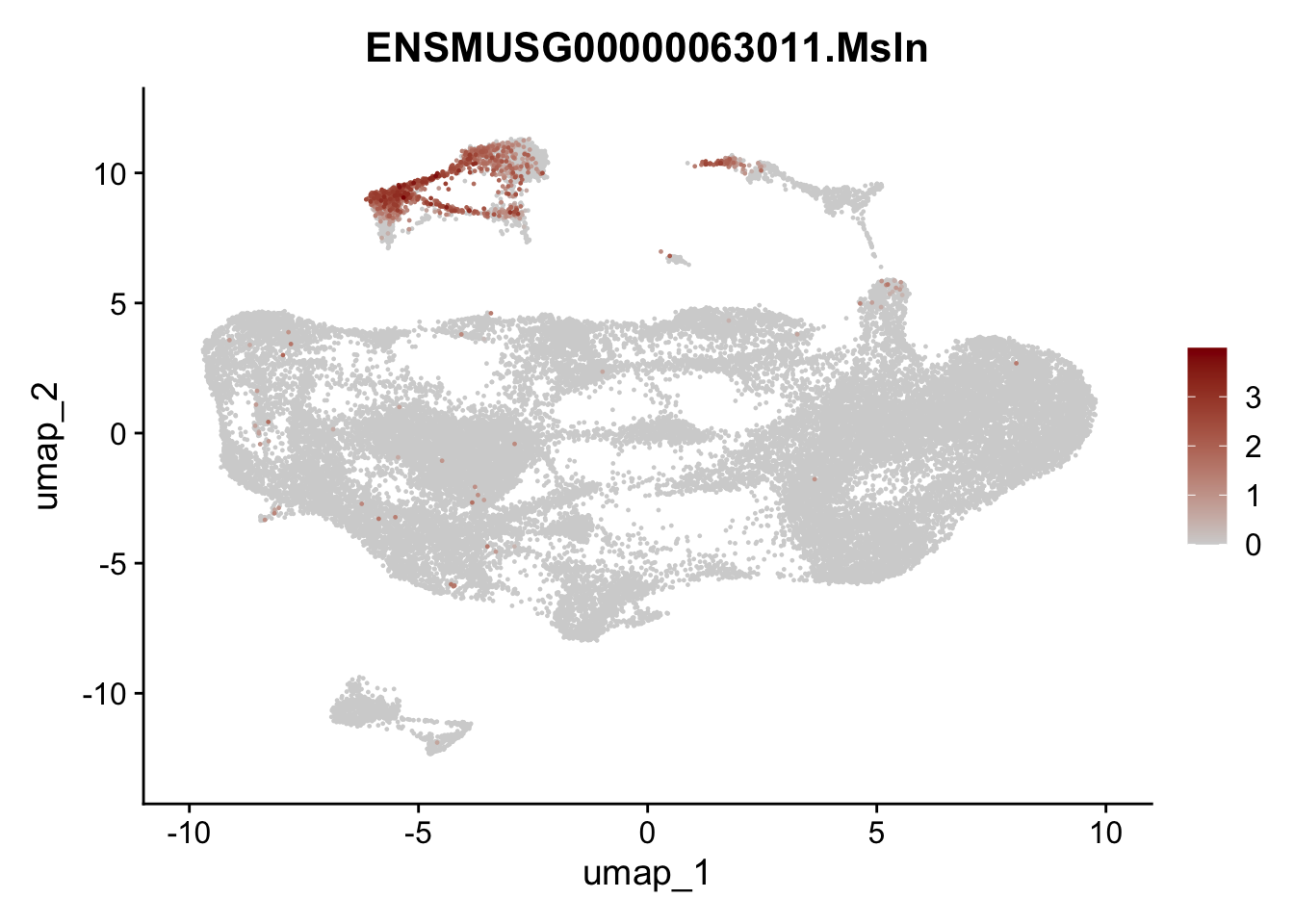
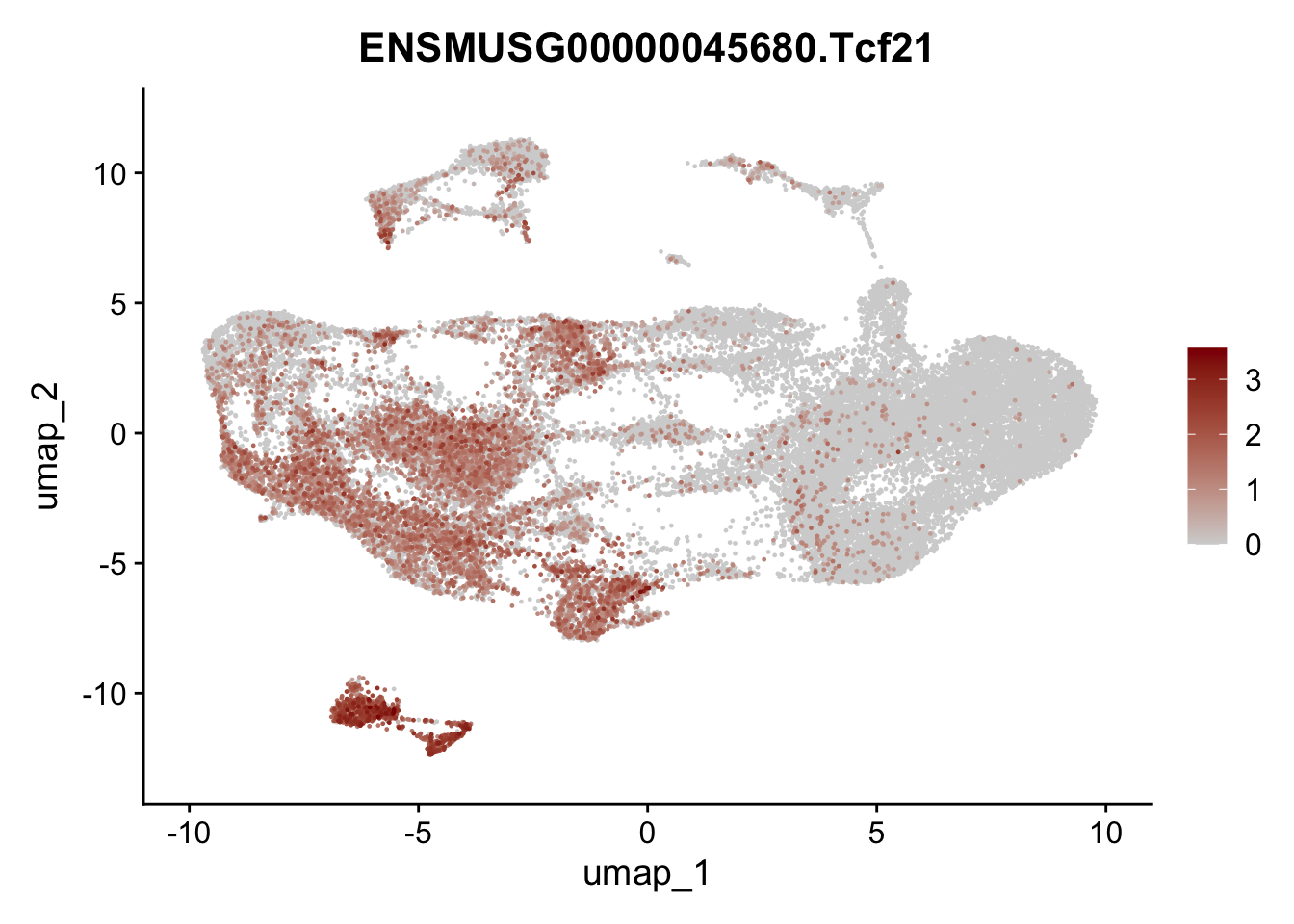
filter
## filter out Ptprc+ cells (cluster #12),
## and pancreatic cells (#15),
## and mesothelial cells (cluster#8)
## and epithelial/neuronal cells (#11)
## and proliferating cells (cluster #5 and #7)
table(seuratmLN$RNA_snn_res.0.4)
seuratF <- subset(seuratmLN, RNA_snn_res.0.4 %in% c("12", "15", "8", "11", "5", "7"), invert = TRUE)
table(seuratF$RNA_snn_res.0.4)
seuratmLNf3 <- seuratF
remove(seuratF)
table(seuratmLNf3$orig.ident)rerun
## rerun seurat
seuratmLNf3 <- NormalizeData (object = seuratmLNf3)
seuratmLNf3 <- FindVariableFeatures(object = seuratmLNf3)
seuratmLNf3 <- ScaleData(object = seuratmLNf3, verbose = TRUE)
seuratmLNf3 <- RunPCA(object=seuratmLNf3, npcs = 30, verbose = FALSE)
seuratmLNf3 <- RunTSNE(object=seuratmLNf3, reduction="pca", dims = 1:20)
seuratmLNf3 <- RunUMAP(object=seuratmLNf3, reduction="pca", dims = 1:20)
seuratmLNf3 <- FindNeighbors(object = seuratmLNf3, reduction = "pca", dims= 1:20)
res <- c(0.25, 0.6, 0.8, 0.4)
for (i in 1:length(res)) {
seuratmLNf3 <- FindClusters(object = seuratmLNf3, resolution = res[i], random.seed = 1234)
}load object mLN EYFP+ fil
fileNam <- paste0(basedir, "/data/LNmLToRev_EYFP_mLNf3_seurat.rds")
seuratmLNf3 <- readRDS(fileNam)
table(seuratmLNf3$dataset)
380131_04-4_20250224_Cxcl13EYFP_P7_mLN_YFPpos 380131_05-5_20250224_Cxcl13EYFP_P7_mLN_YFPneg
3922 12
380131_07-7_20250225_Cxcl13EYFP_E18_mLN_YFPpos 380131_08-8_20250225_Cxcl13EYFP_E18_mLN_YFPneg
2392 5
380131_12-12_20250305_Mu_Cxcl13EYFP_Adult_mLN_FRC 382581_02-2_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPpos
3798 3617
382581_03-3_20250311_Mu_Cxcl13EYFP_E18_mLN_YFPneg 382581_06-6_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPpos
15 4274
382581_07-7_20250319_Mu_Cxcl13EYFP_P7_mLN_YFPneg 382581_09-9_20250320_Mu_Cxcl13EYFP_Adult_mLN_FRC
11 2711
382581_14-14_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib1 382581_15-15_20250402_Mu_Cxcl13EYFP_3wk_mLN_fib2
3223 4289 dimplot
clustering
colPal <- c("#DAF7A6", "#FFC300", "#FF5733", "#C70039", "#900C3F", "#b66e8d",
"#61a4ba", "#6178ba", "#54a87f", "#25328a",
"#b6856e", "#0073C2FF", "#EFC000FF", "#868686FF", "#CD534CFF",
"#7AA6DCFF", "#003C67FF", "#8F7700FF", "#3B3B3BFF", "#A73030FF",
"#4A6990FF")[1:length(unique(seuratmLNf3$RNA_snn_res.0.4))]
names(colPal) <- unique(seuratmLNf3$RNA_snn_res.0.4)
DimPlot(seuratmLNf3, reduction = "umap", group.by = "RNA_snn_res.0.4",
cols = colPal, label = TRUE)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")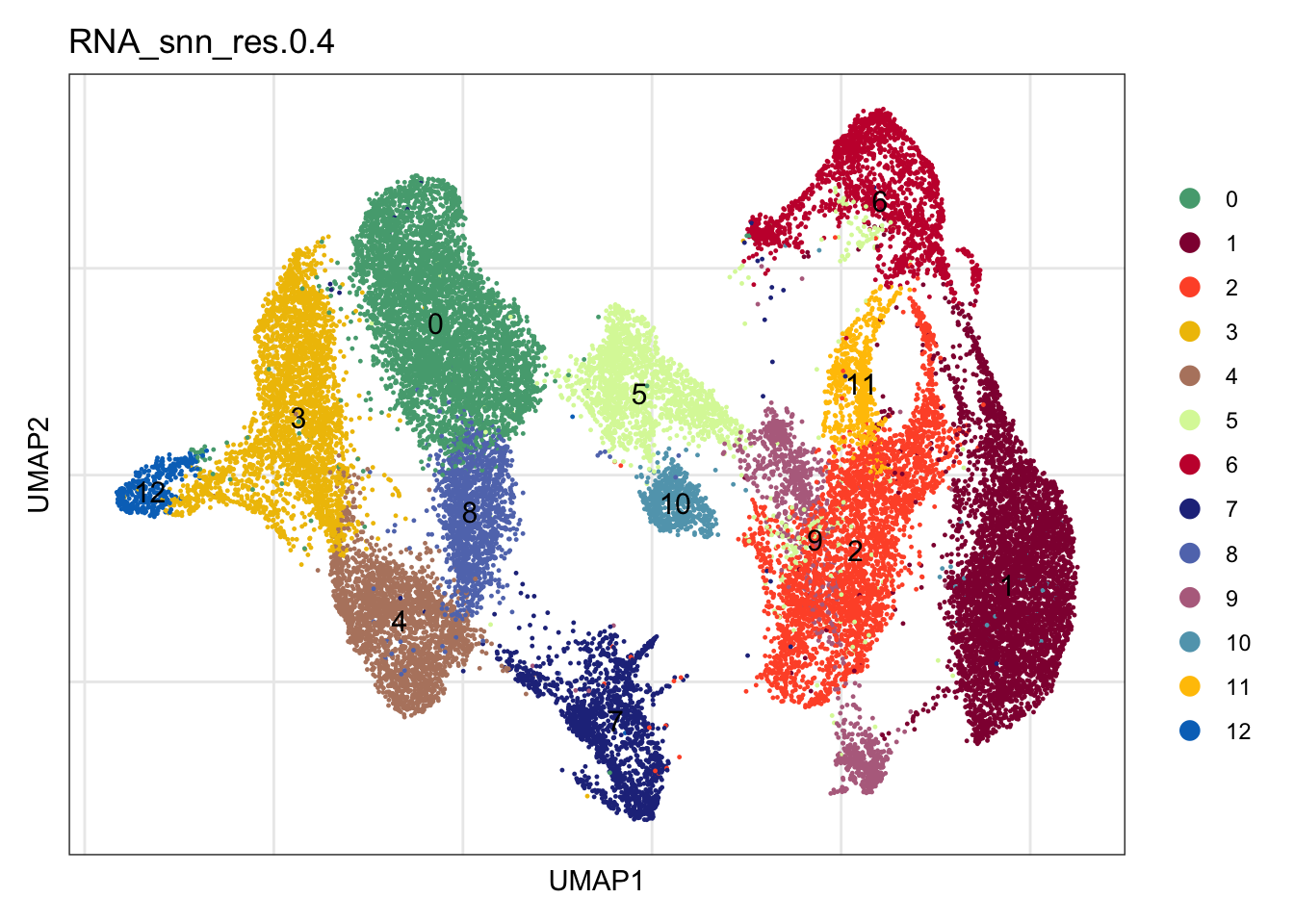
timepoint
DimPlot(seuratmLNf3, reduction = "umap", group.by = "timepoint",
cols = coltimepoint)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")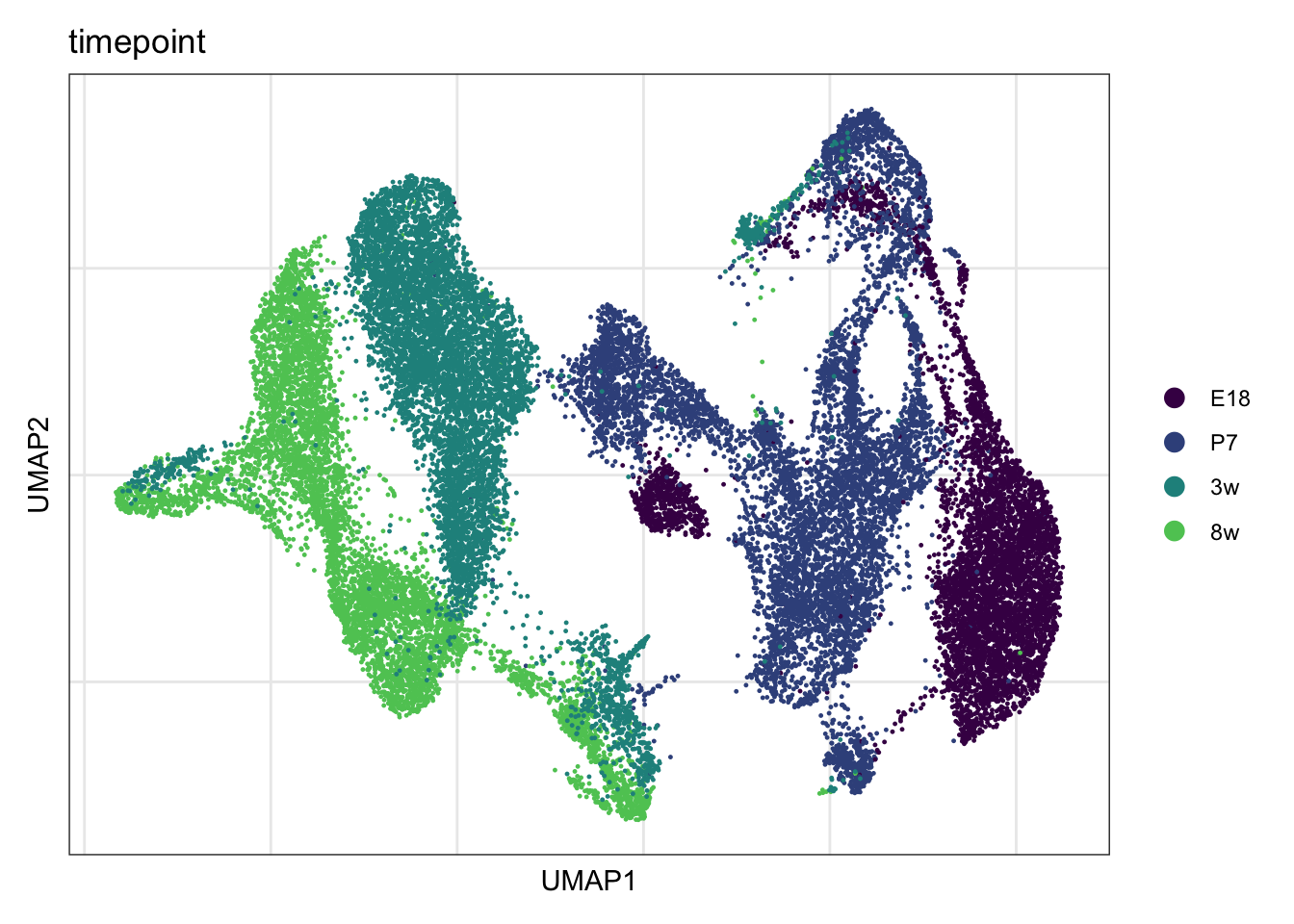
label transfer
fileNam <- paste0(basedir, "/data/LNmLToRev_adultonly_seurat.integrated.rds")
seuratLab <- readRDS(fileNam)
seuratLab <- subset(seuratLab, location=="mLN")
seuratLab <- subset(seuratLab, EYFP=="pos")
table(seuratLab$label)
actMedRC FDC/MRC MedRC MedRC/IFRC Pi16+RC PRC TBRC TRC VSMC
1047 692 275 1259 262 673 1291 913 47 dimplot label
labCells <- data.frame(label=seuratLab$label) %>% rownames_to_column(., "cell")
allCell <- data.frame(cell=colnames(seuratmLNf3)) %>%
left_join(., labCells, by= "cell")
allCell$label[which(is.na(allCell$label))] <- "unassigned"
seuratmLNf3$label <- allCell$label
table(seuratmLNf3$timepoint)
E18 P7 3w 8w
6029 8219 7512 6509 table(seuratmLNf3$label)
actMedRC FDC/MRC MedRC MedRC/IFRC Pi16+RC PRC TBRC TRC unassigned
1047 689 274 1259 262 673 1291 913 21814
VSMC
47 colLab <- c("#42a071", "#900C3F","#b66e8d", "#61a4ba", "#424671", "#003C67FF",
"#e3953d", "#714542", "#b6856e", "#a4a4a4")
names(colLab) <- c("FDC/MRC", "TRC", "TBRC", "MedRC/IFRC", "MedRC" , "actMedRC",
"PRC", "Pi16+RC", "VSMC", "unassigned")
DimPlot(seuratmLNf3, reduction = "umap", group.by = "label",
cols = colLab, shuffle=T)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")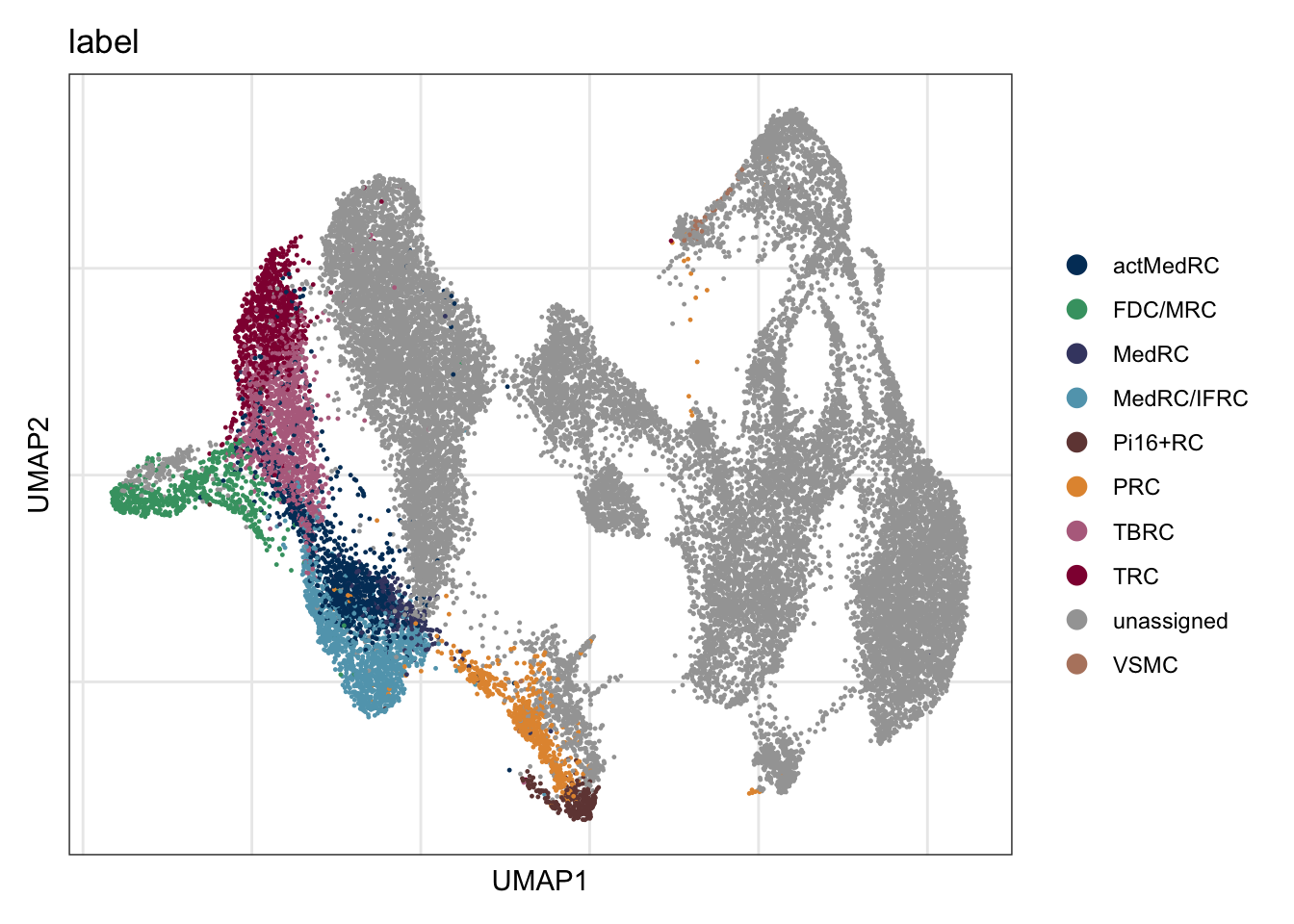
DimPlot(seuratmLNf3, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, order = TRUE)+
theme_void()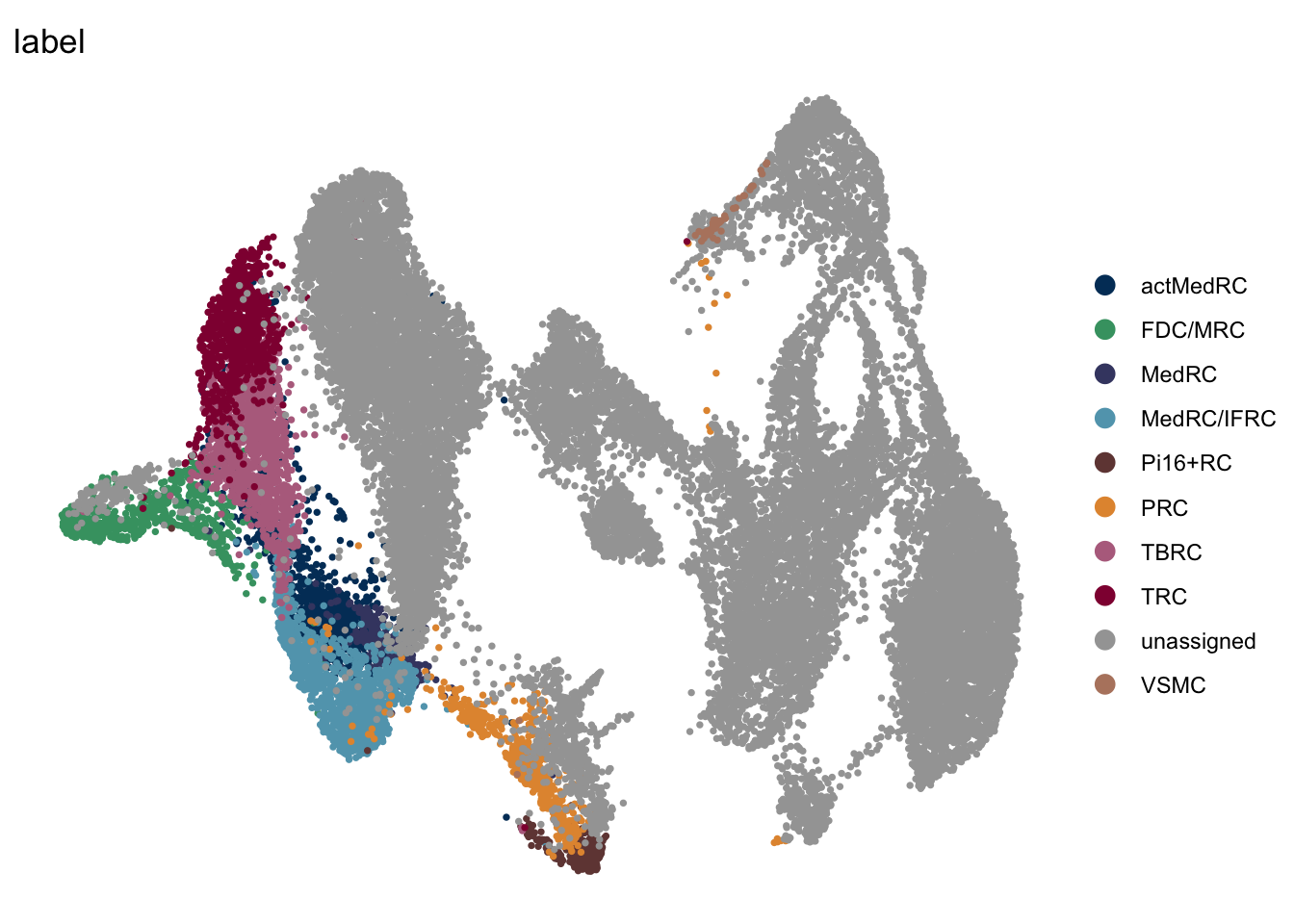
DimPlot(seuratmLNf3, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = FALSE)+
theme_void()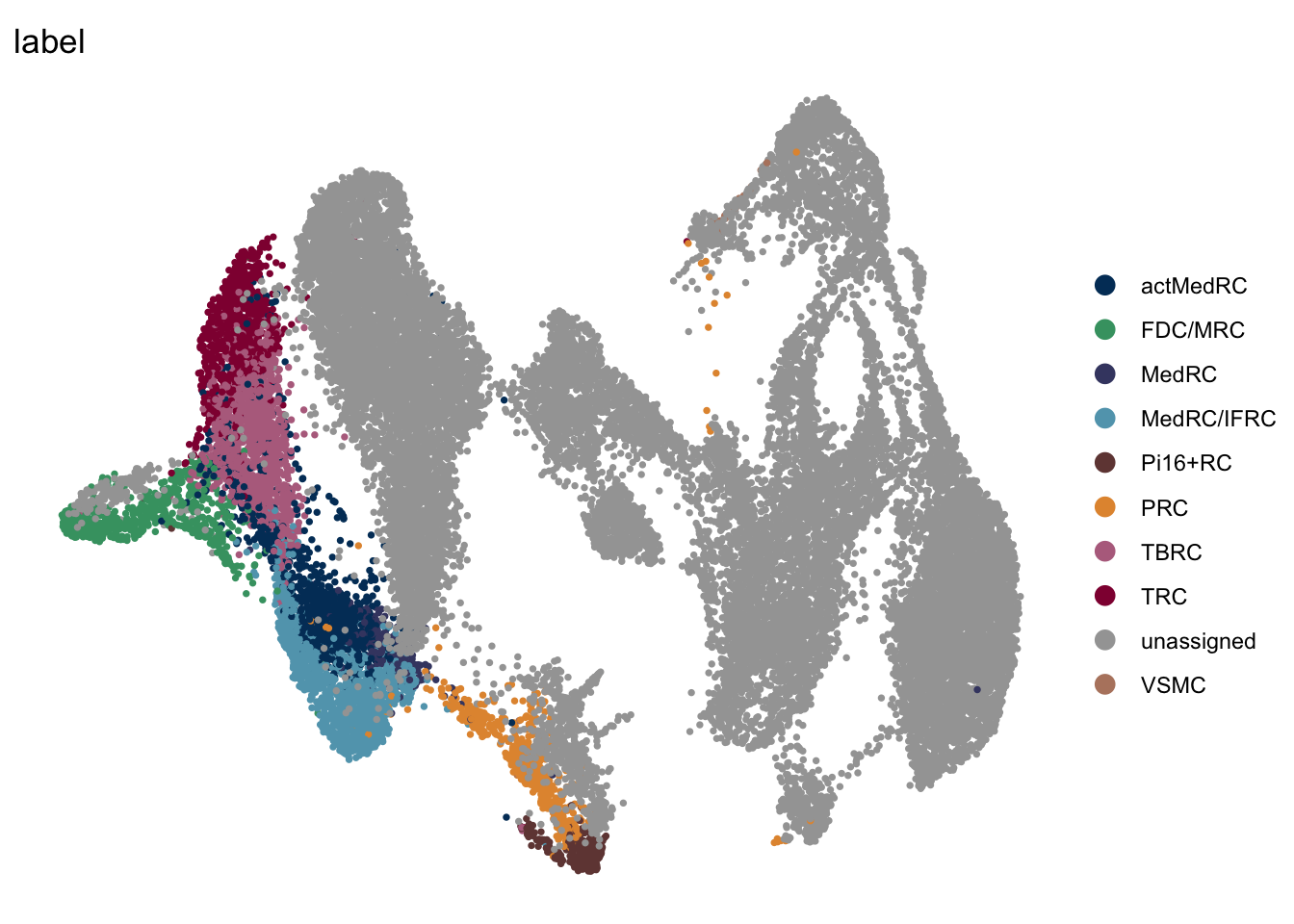
calculate cluster marker genes mLNf3
##cluster marker
Idents(seuratmLNf3) <- seuratmLNf3$RNA_snn_res.0.4
markerGenes <- FindAllMarkers(seuratmLNf3, only.pos=T) %>%
dplyr::filter(p_val_adj < 0.01)features
genes <- data.frame(gene=rownames(seuratmLNf3)) %>%
mutate(geneID=gsub("^.*\\.", "", gene))
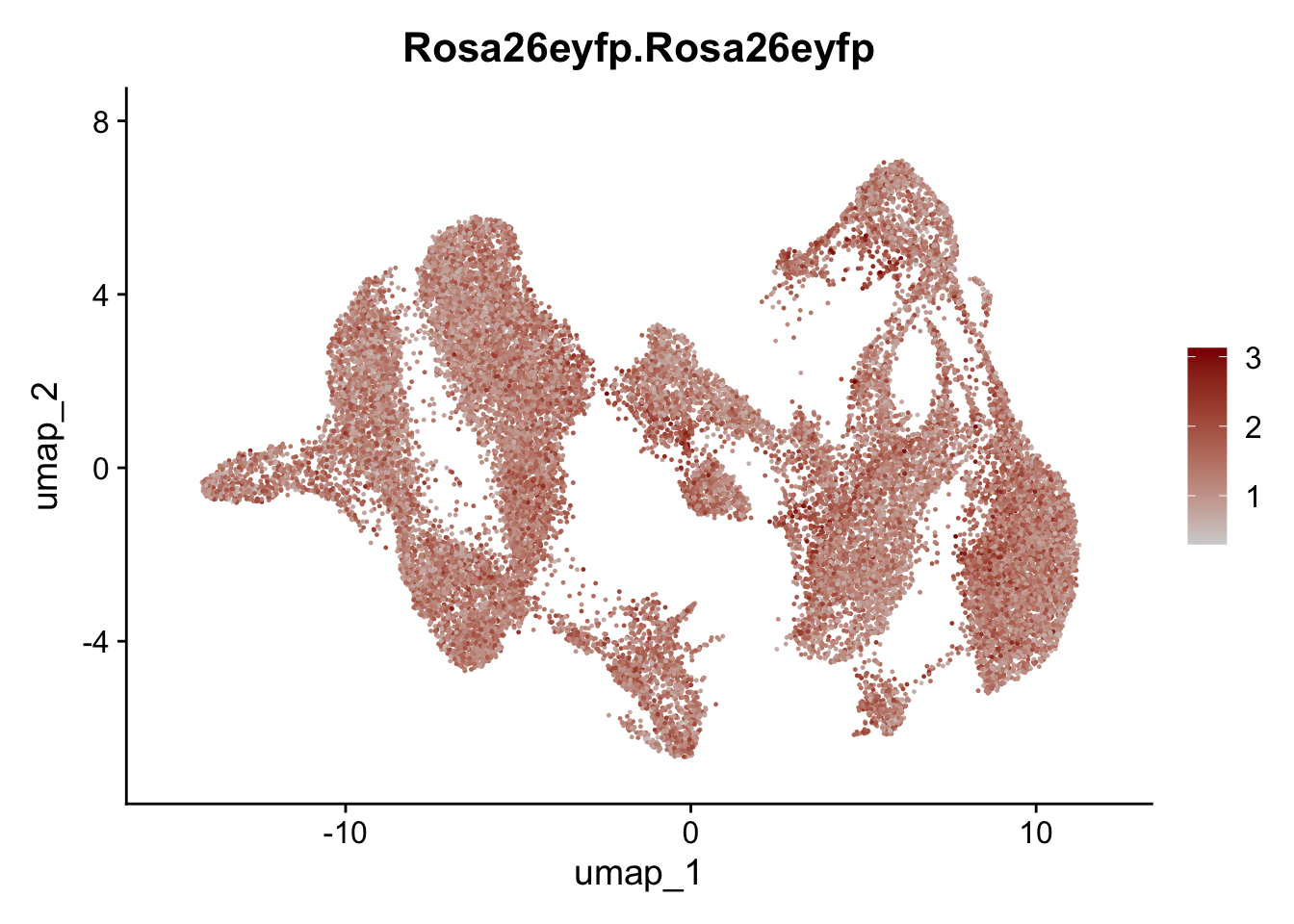
selGenesAll <- data.frame(geneID=c("Rosa26eyfp","tdTomato", "Ccl19", "Ccl21a", "Cxcl13",
"Fbln1", "Col15a1", "Cnn1", "Acta2","Myh11", "Rgs5",
"Cox4i2", "Pi16", "Cd34", "Emp1", "Ogn","Des",
"Fhl2", "Bmp2", "Bmp4", "Grem1", "Grem2", "Bmpr1a", "Bmpr1b", "Bmpr2")) %>%
left_join(., genes, by = "geneID")
pList <- sapply(selGenesAll$gene, function(x){
p <- FeaturePlot(seuratmLNf3, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = F) +
theme(legend.position="right")
plot(p)
})
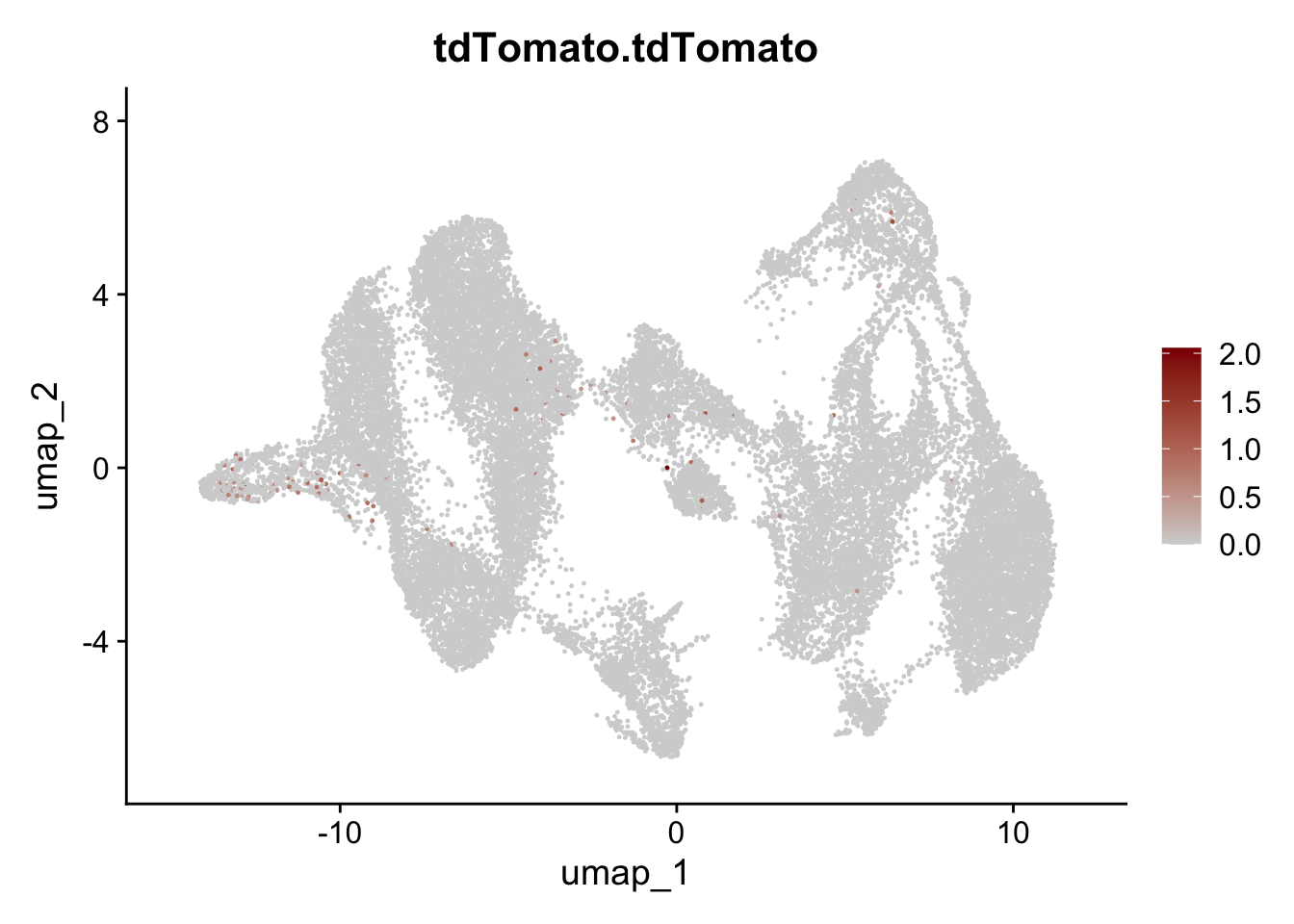
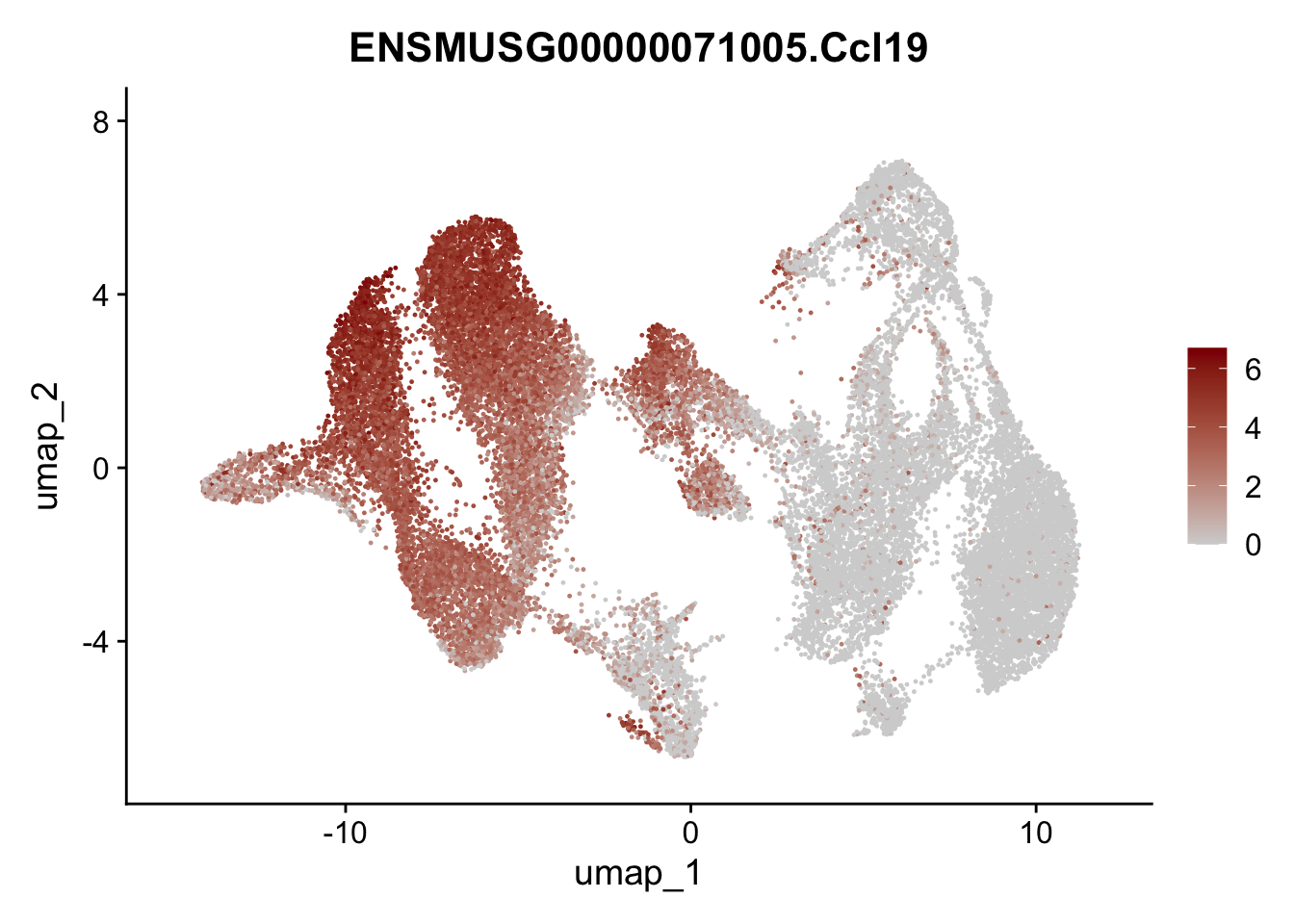
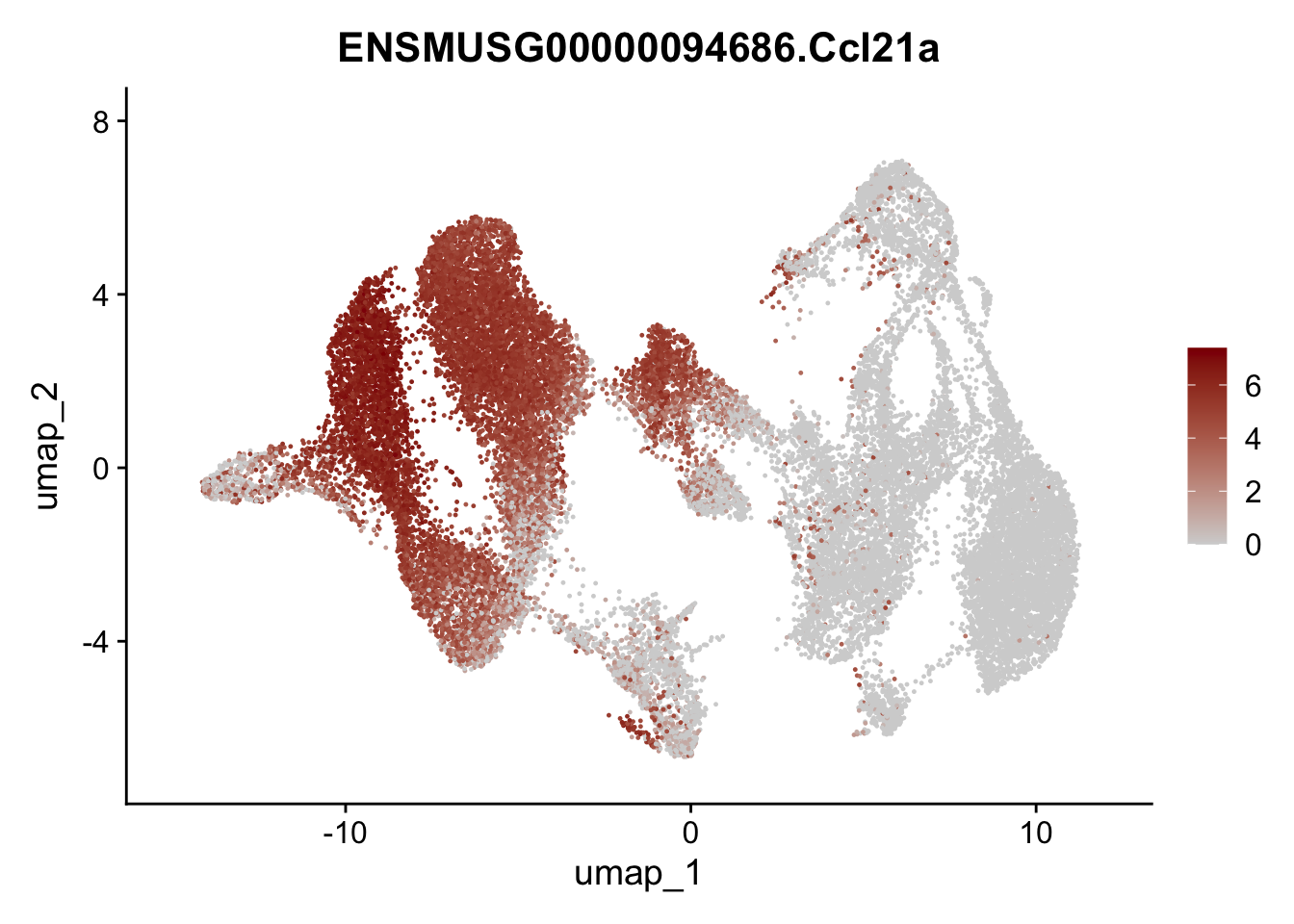
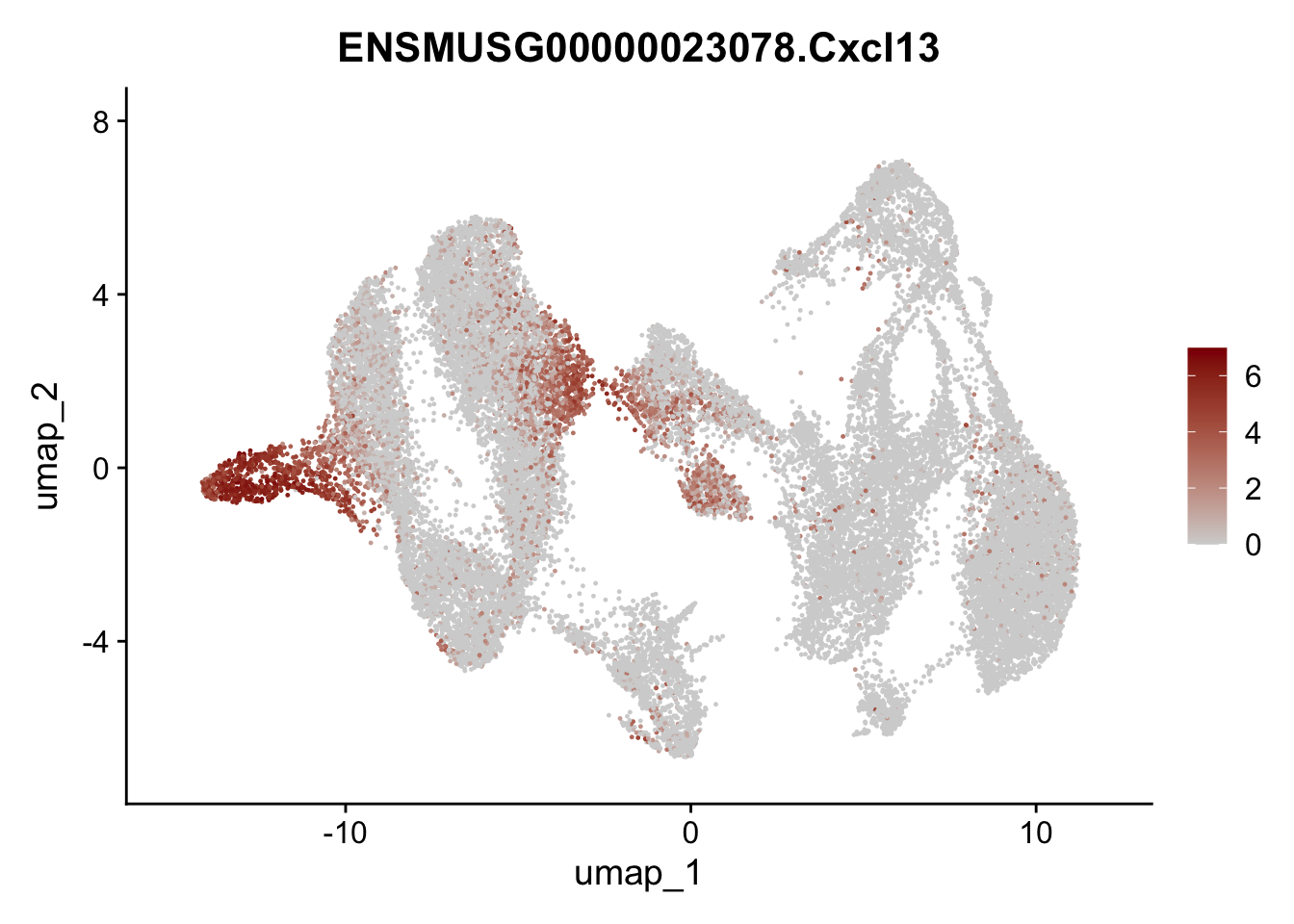
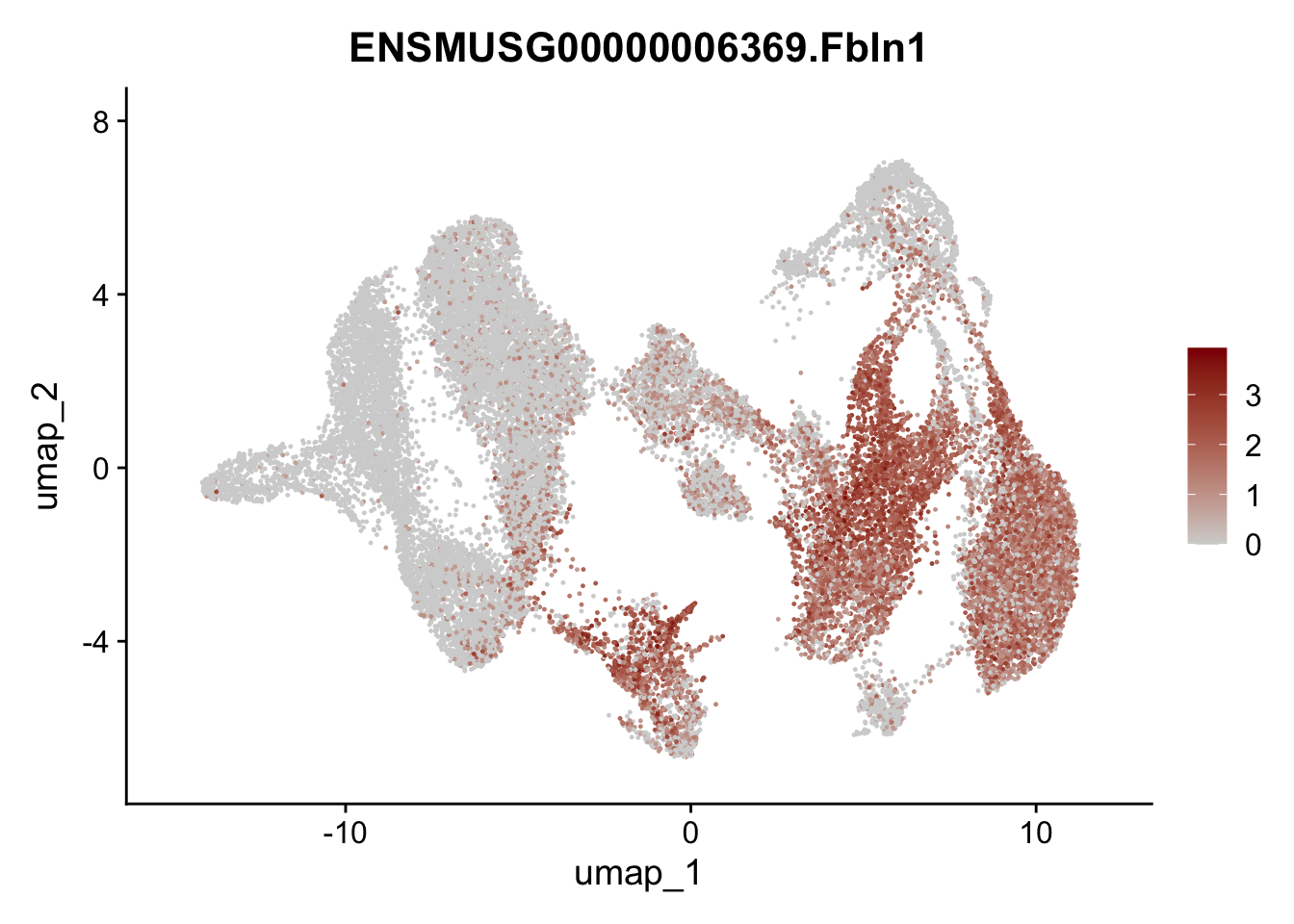
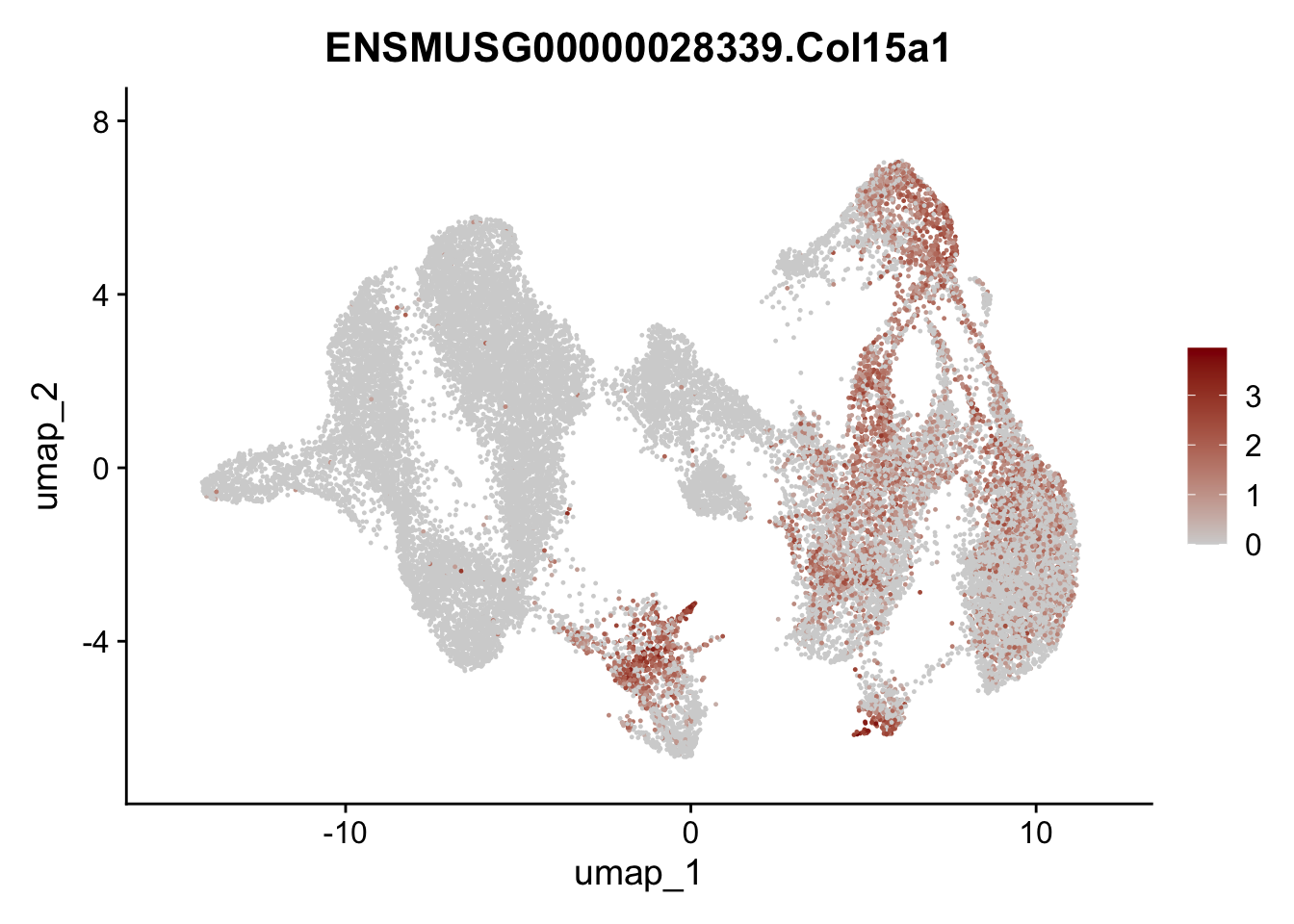
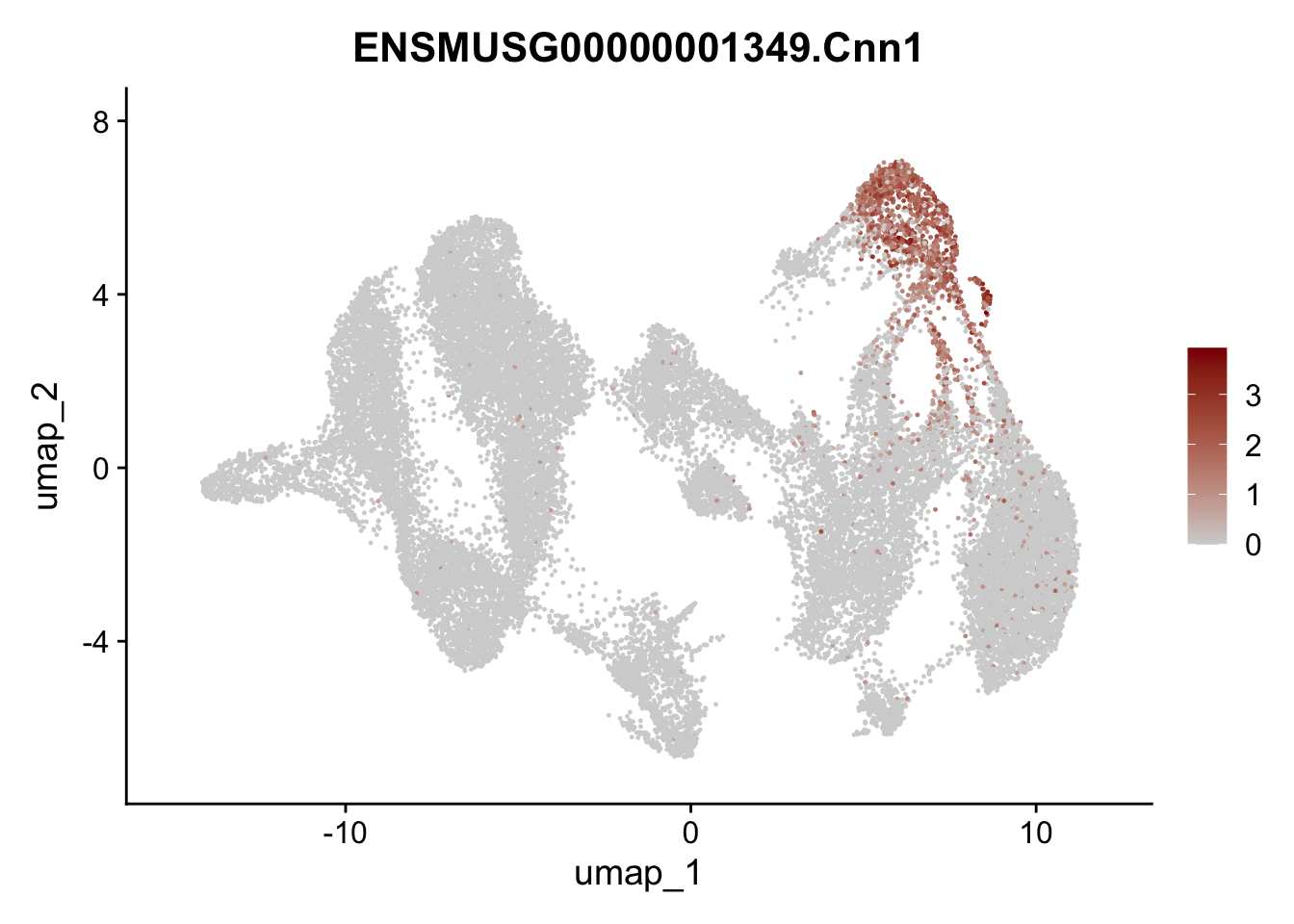
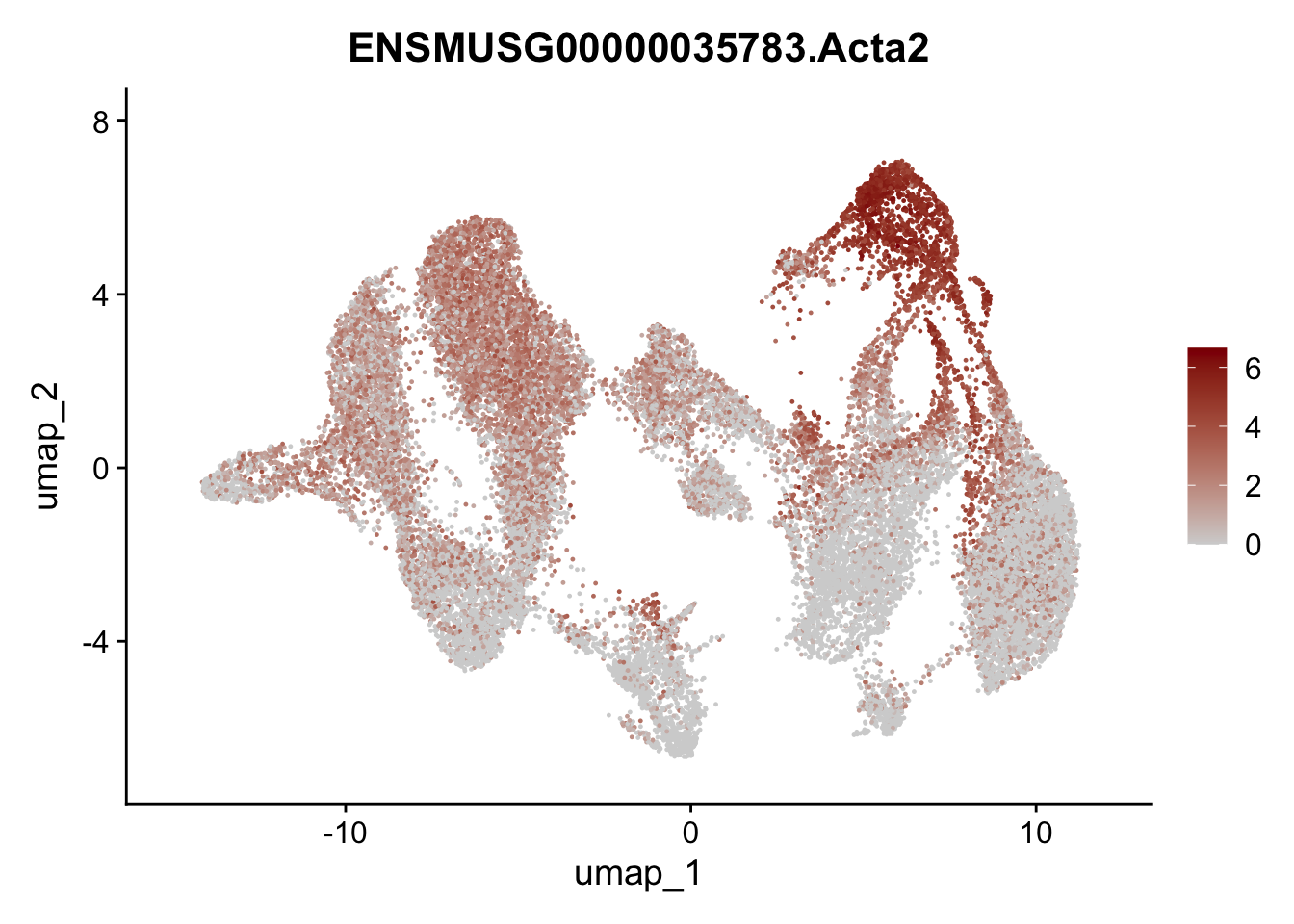
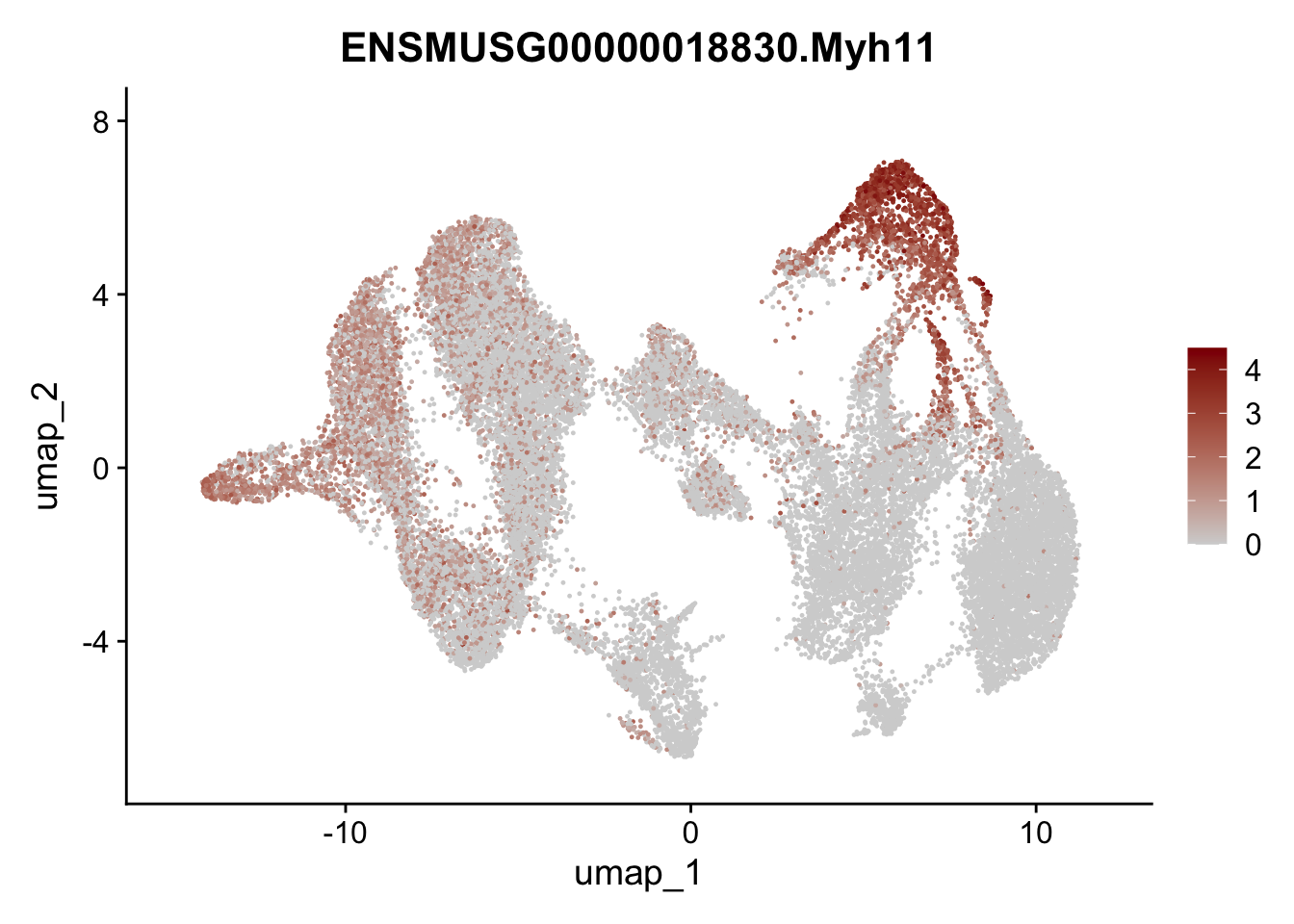
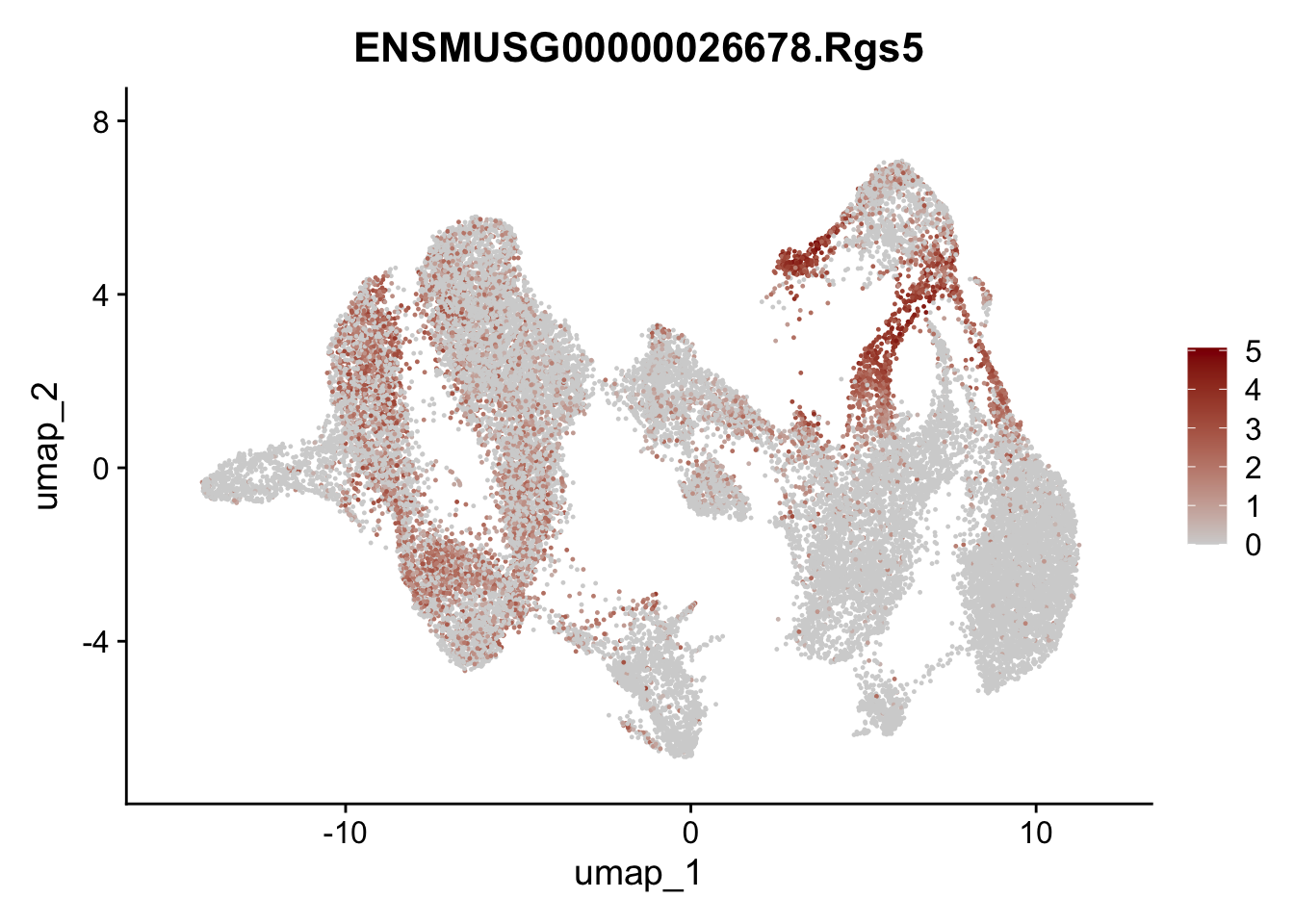
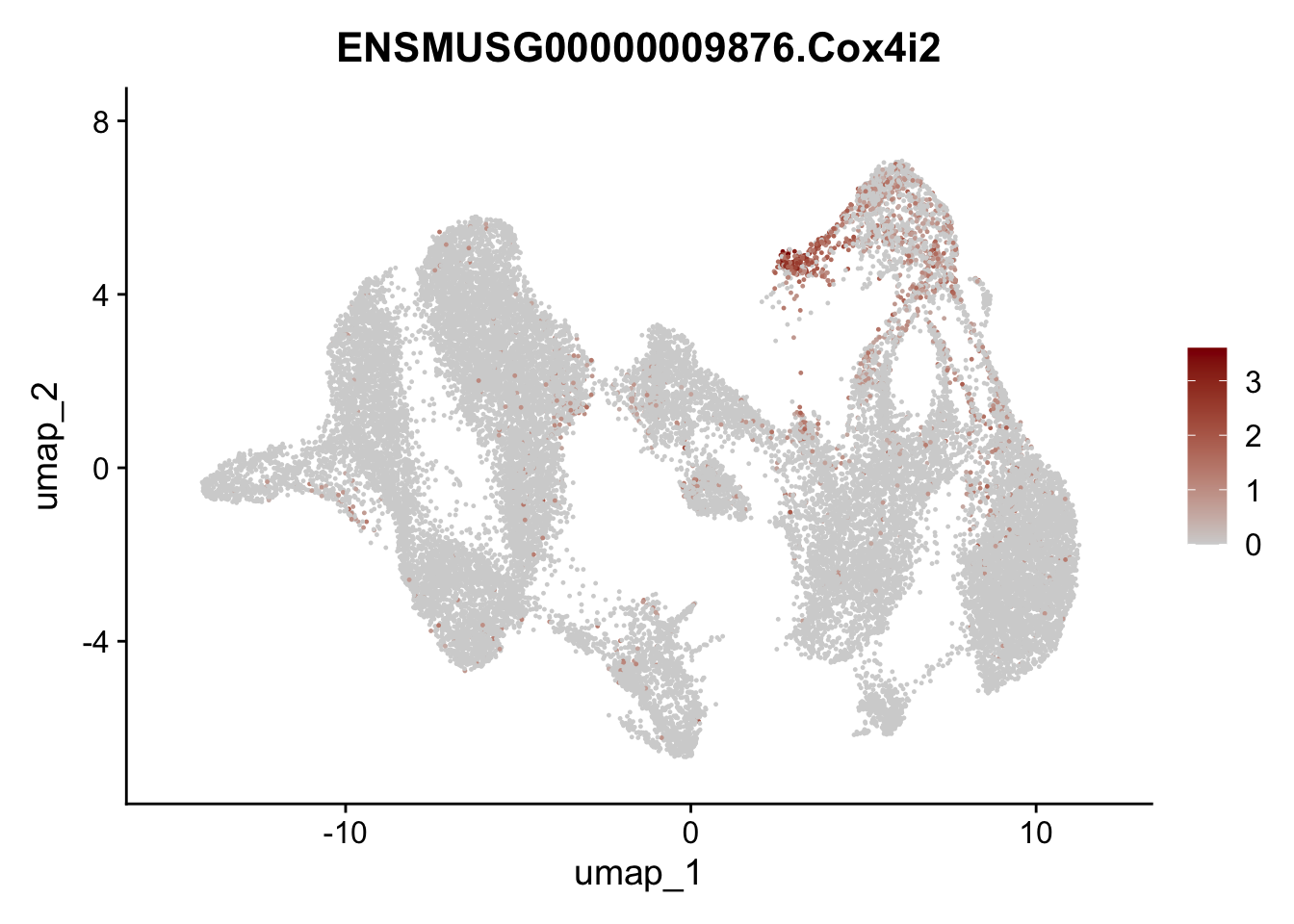
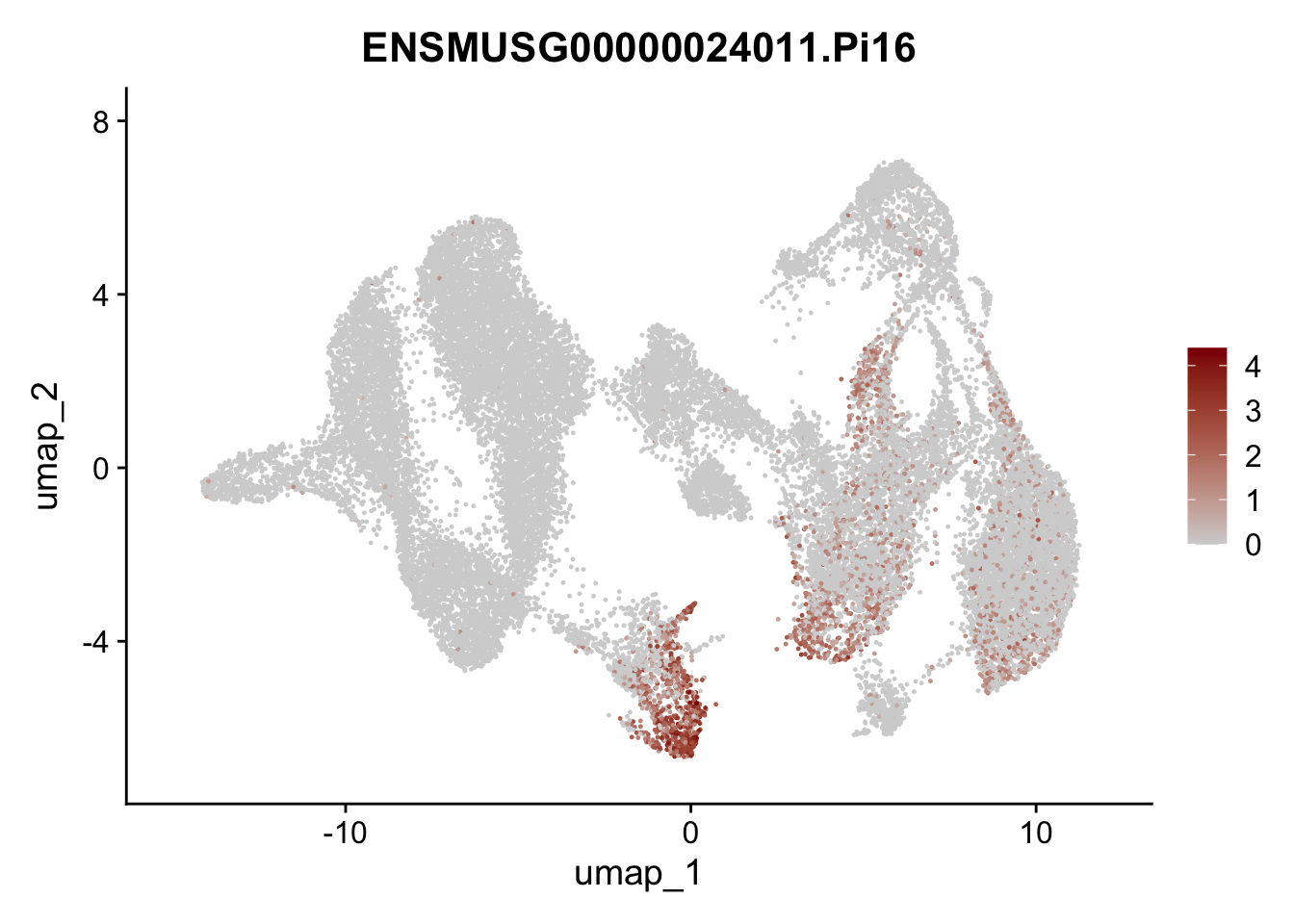
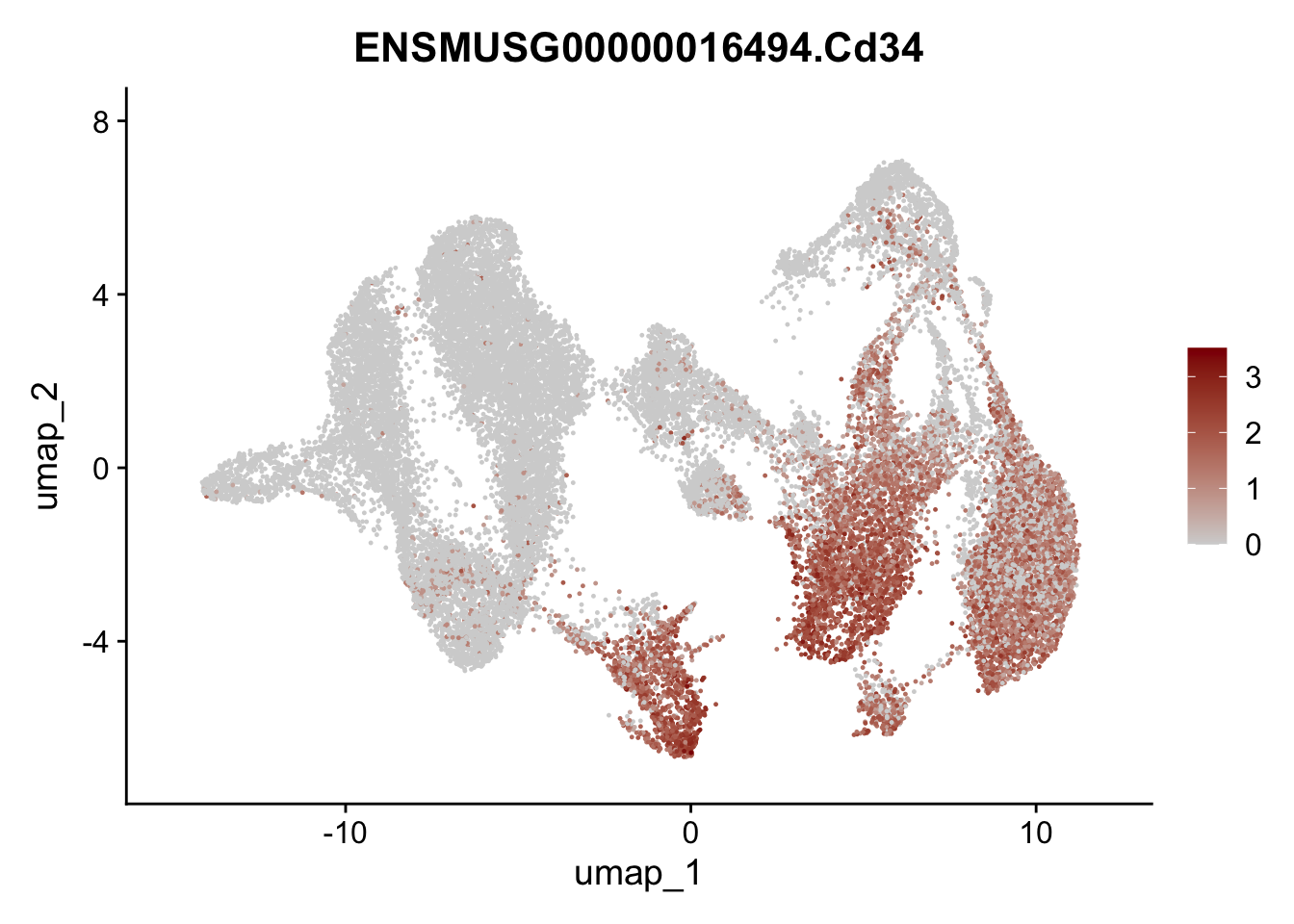
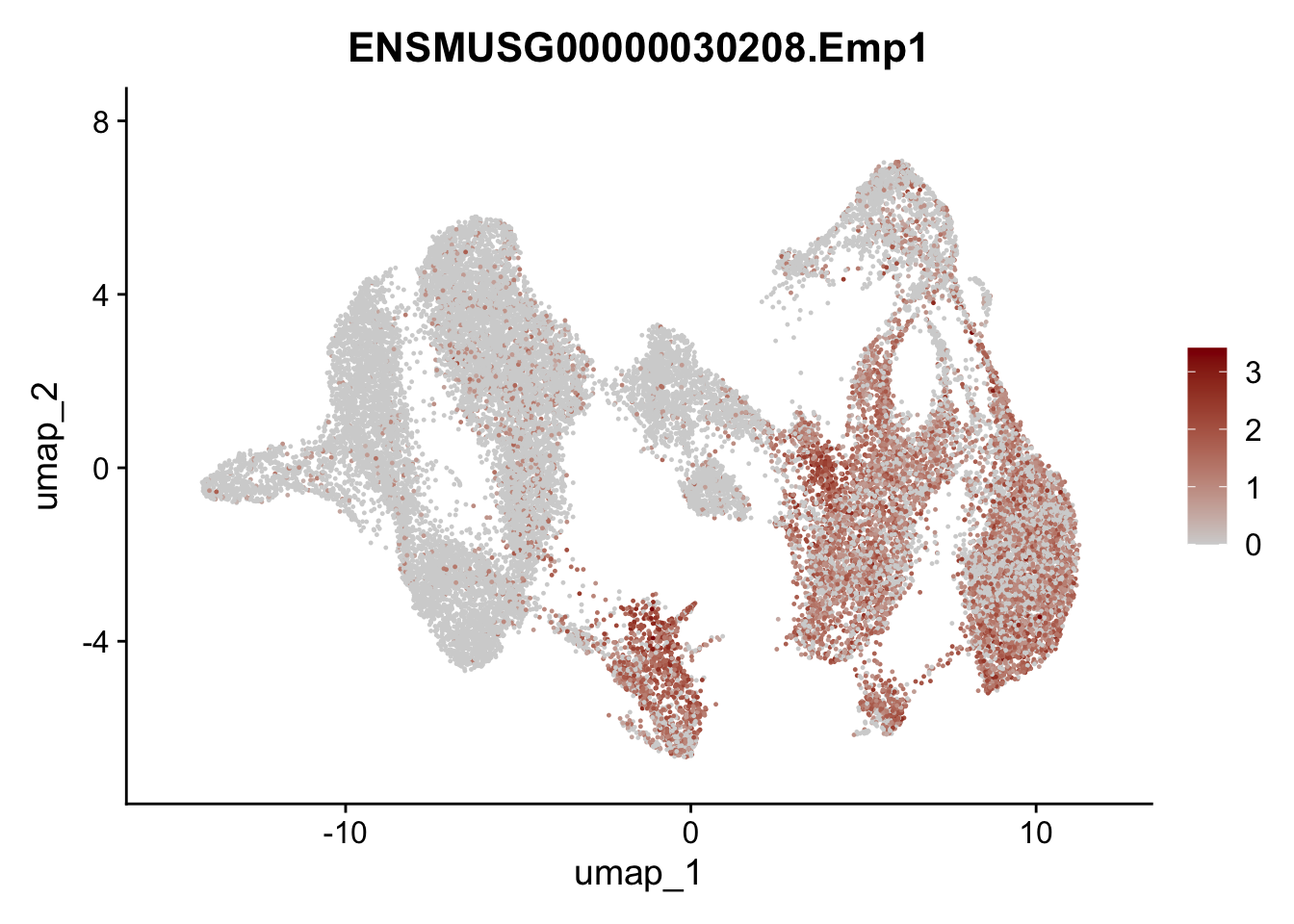
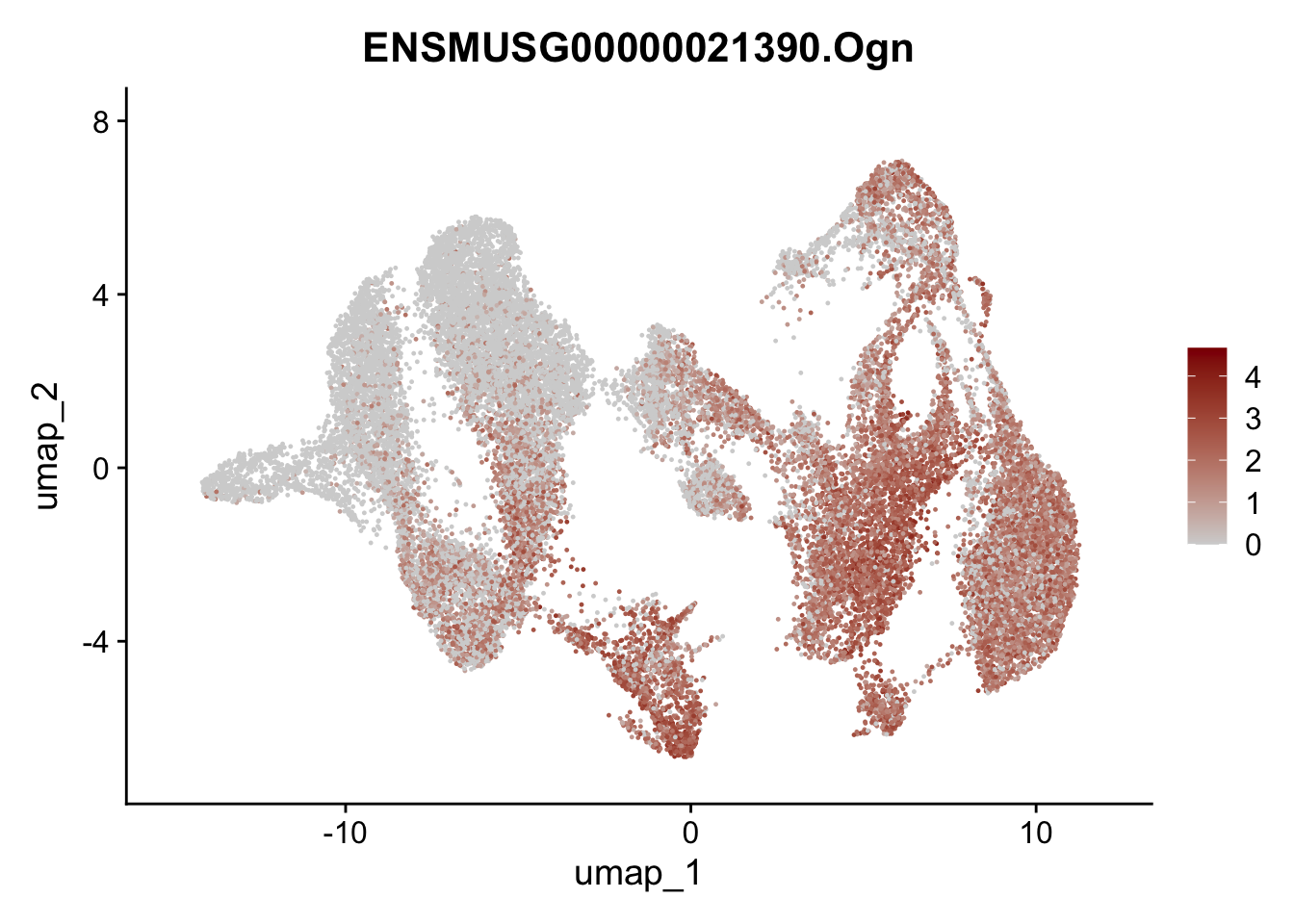
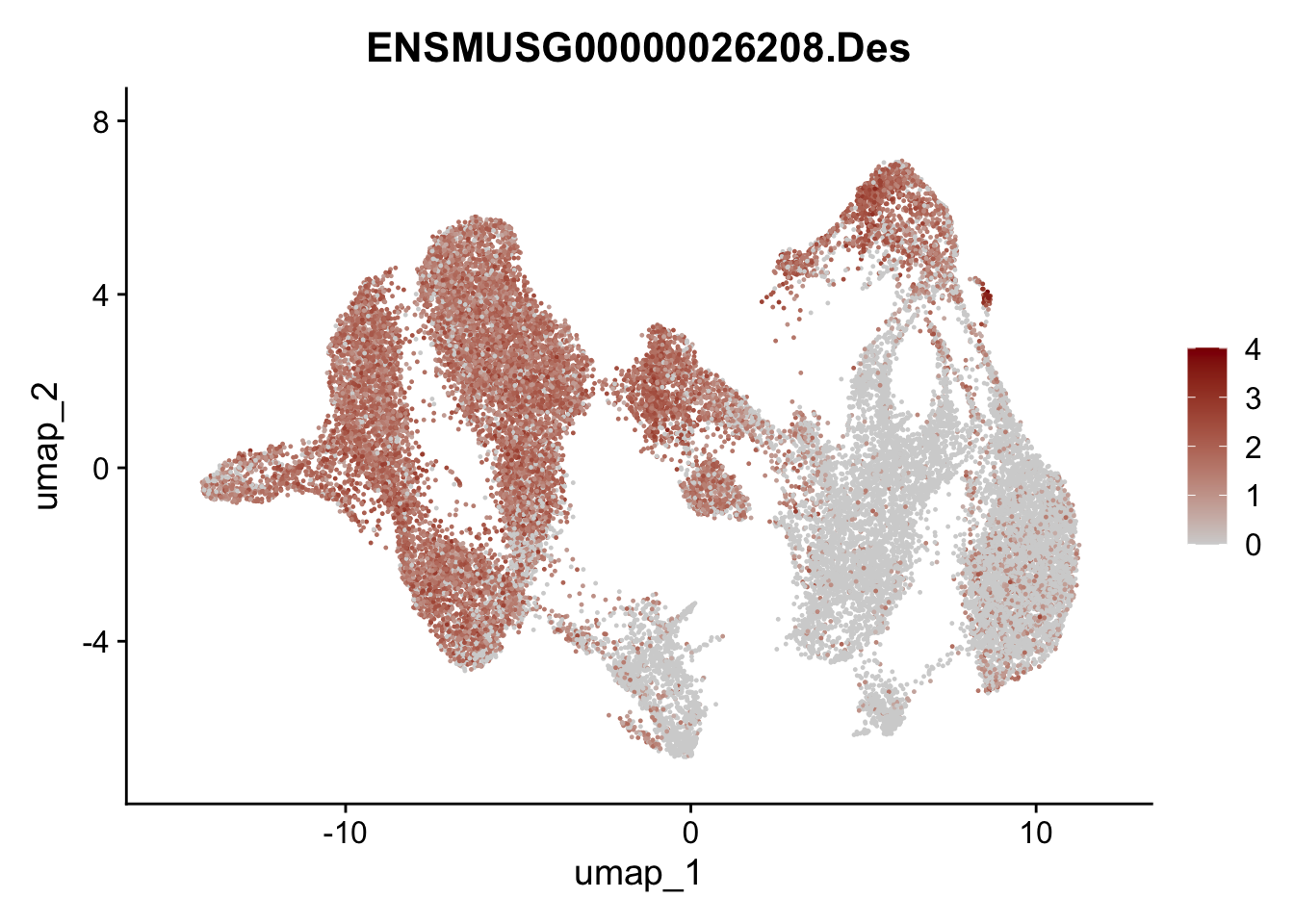
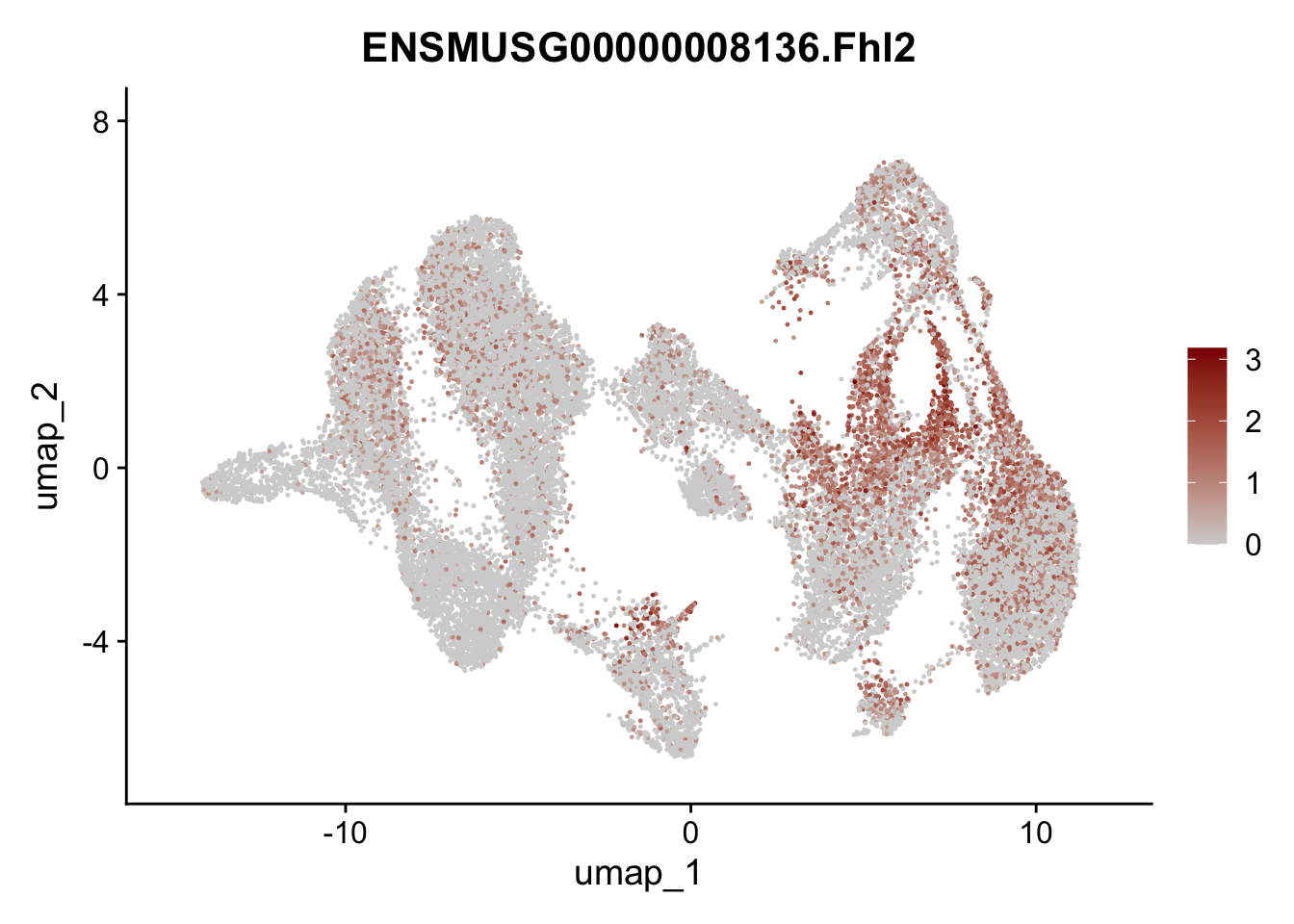
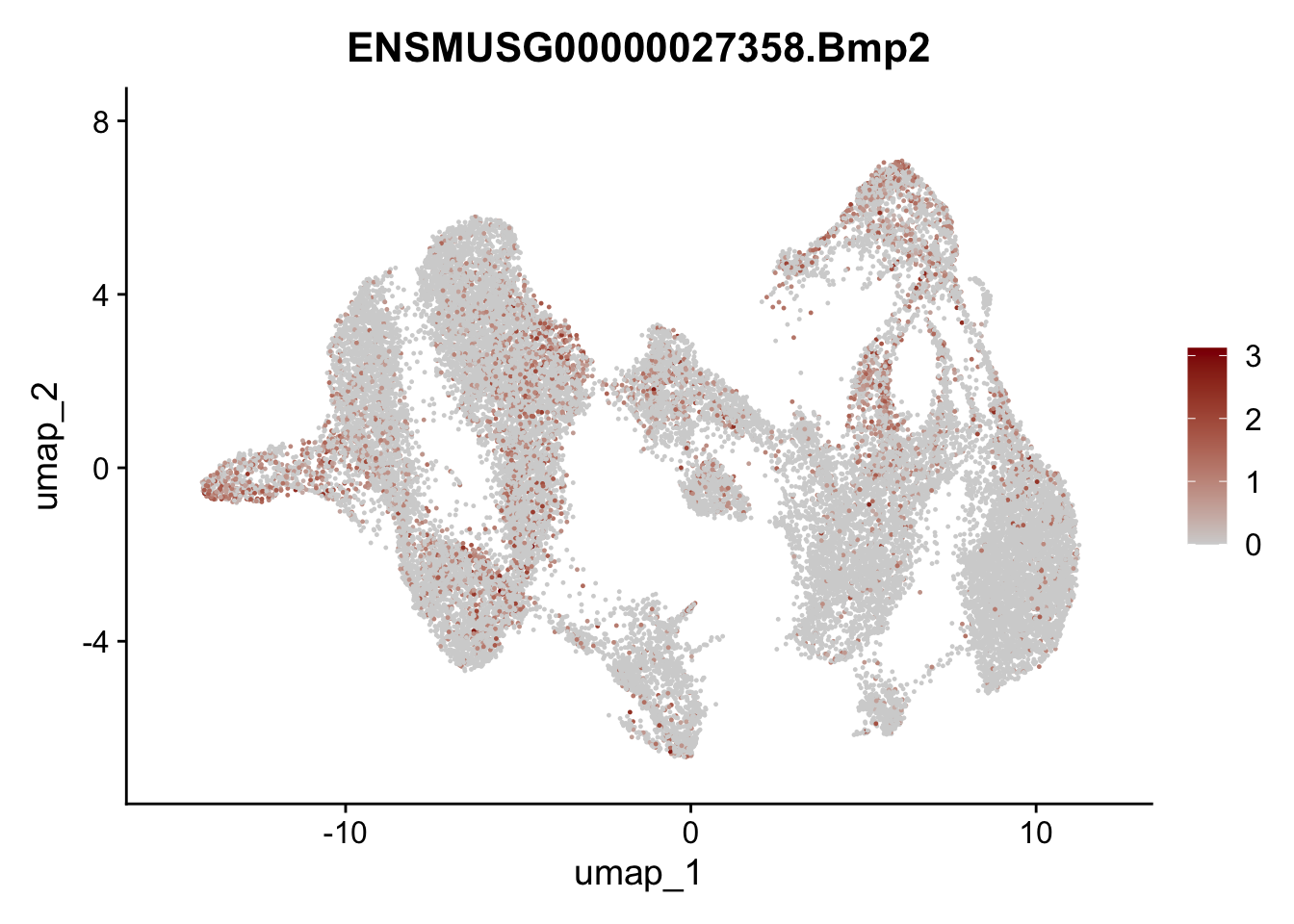
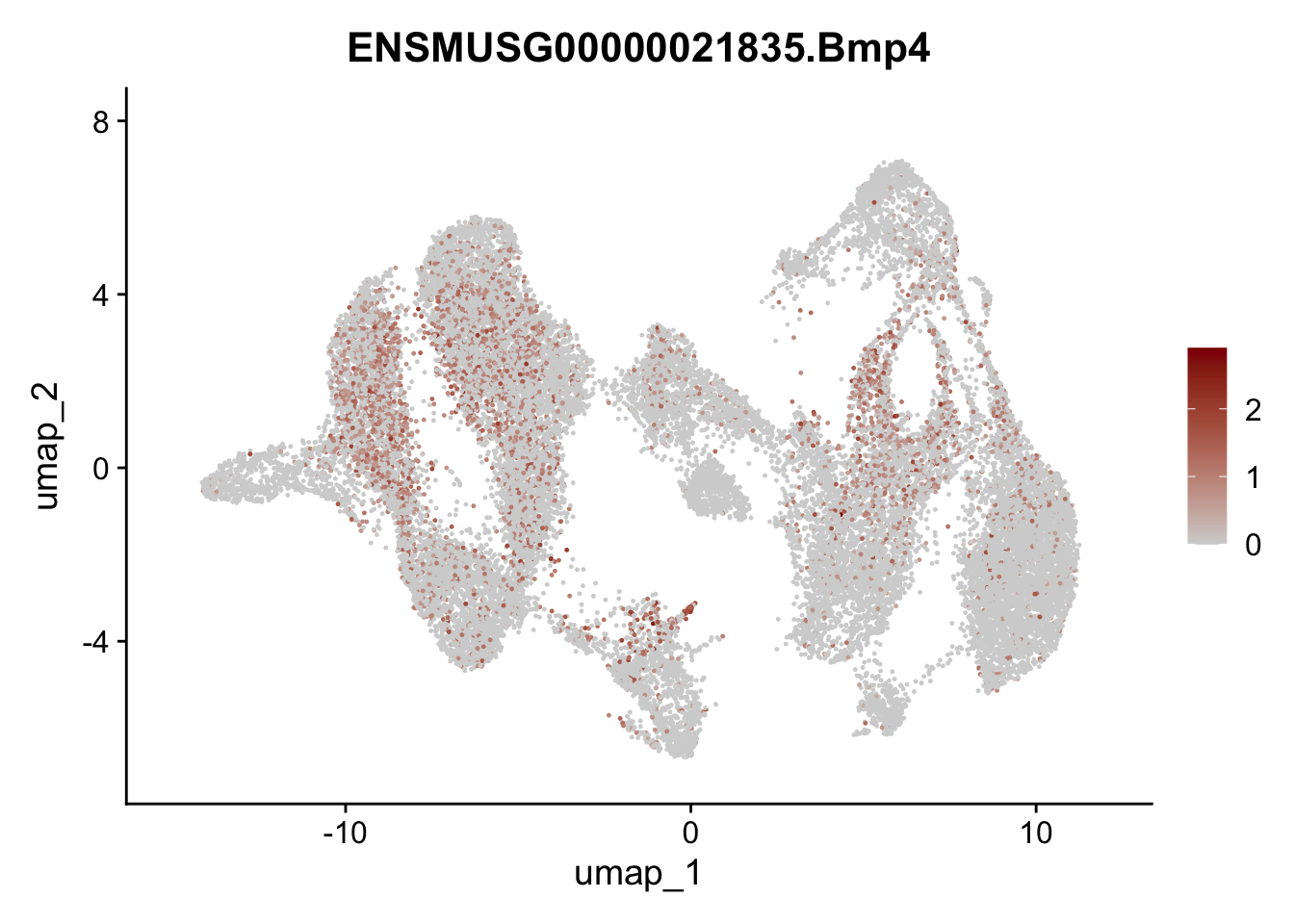
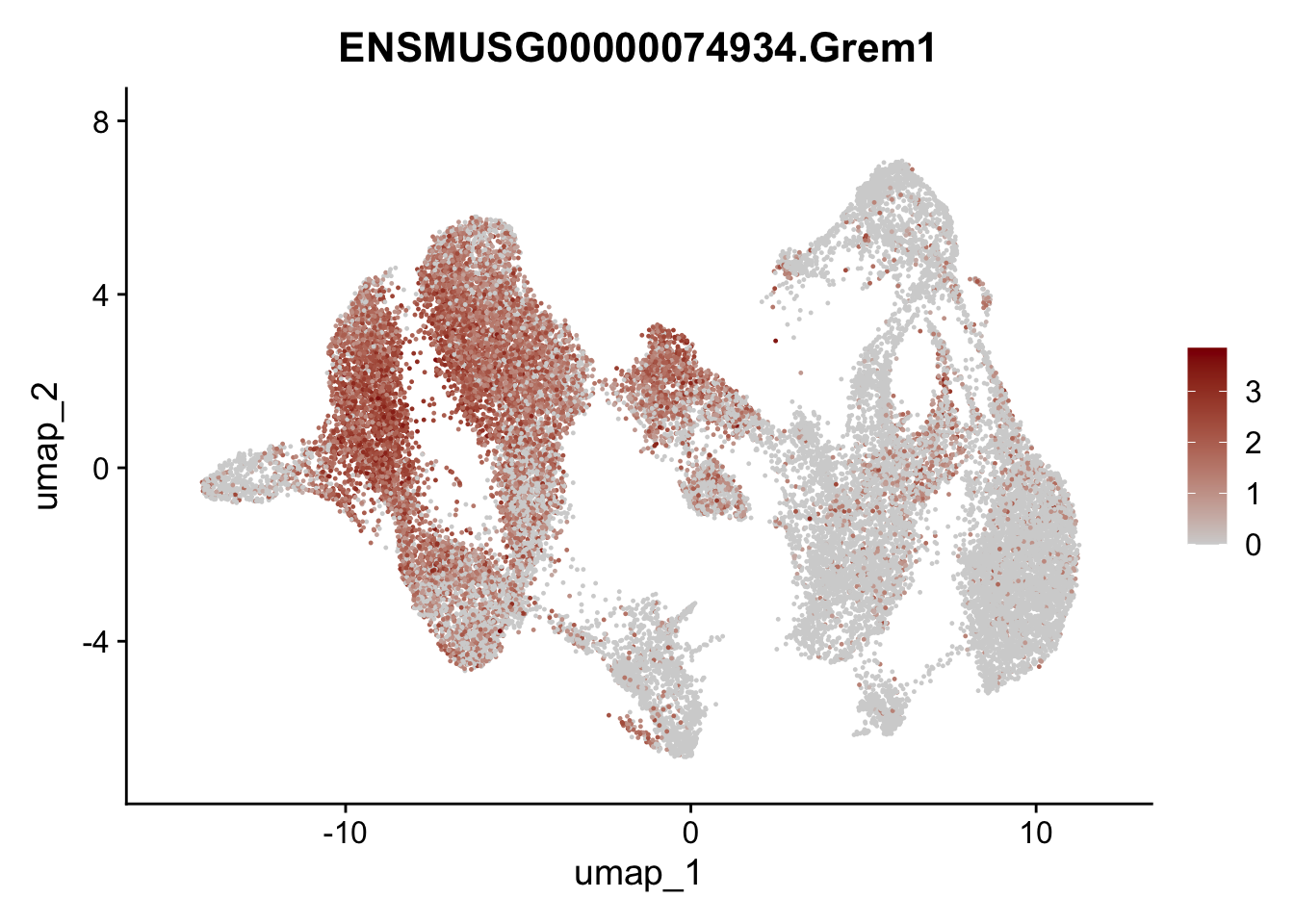
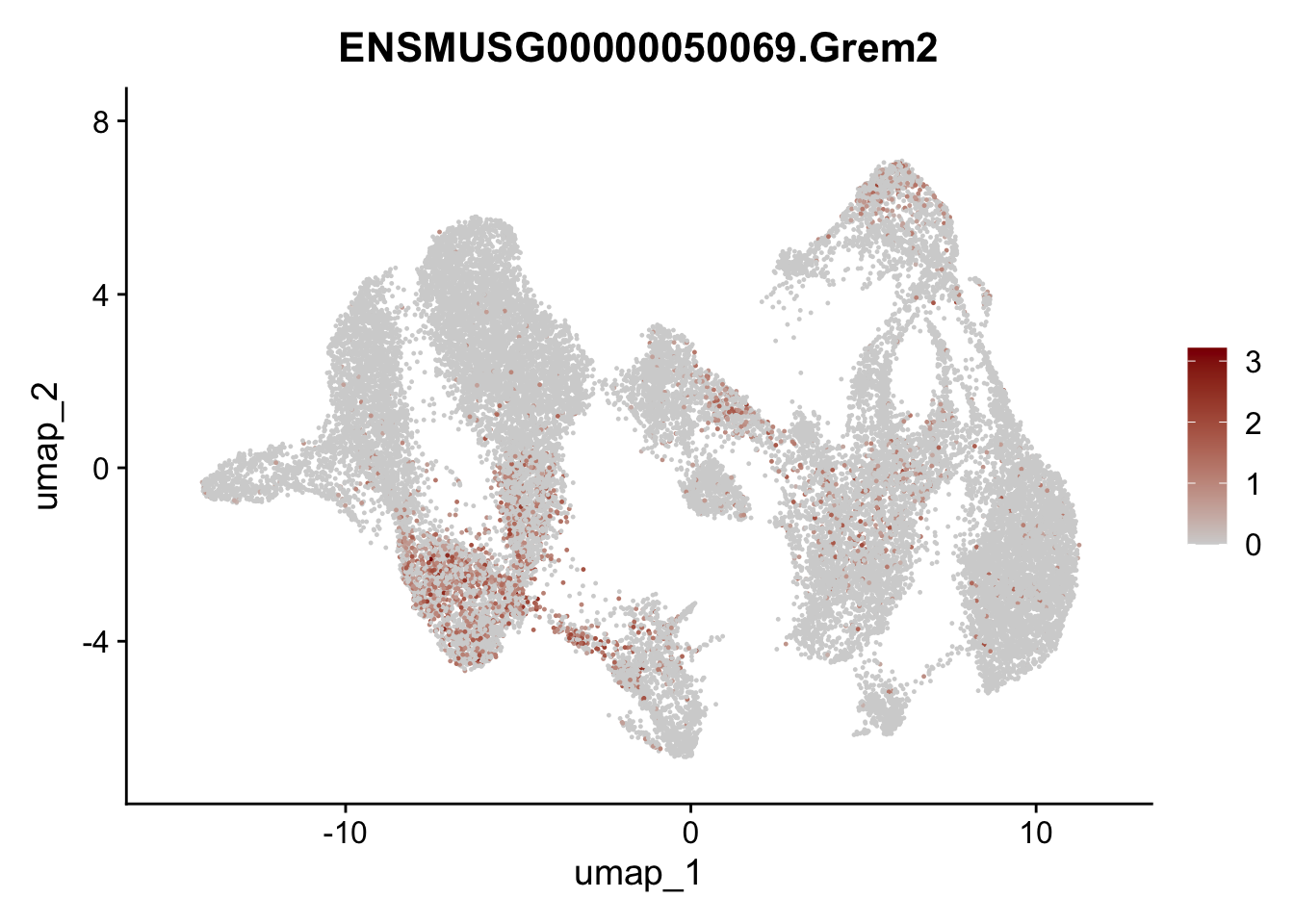
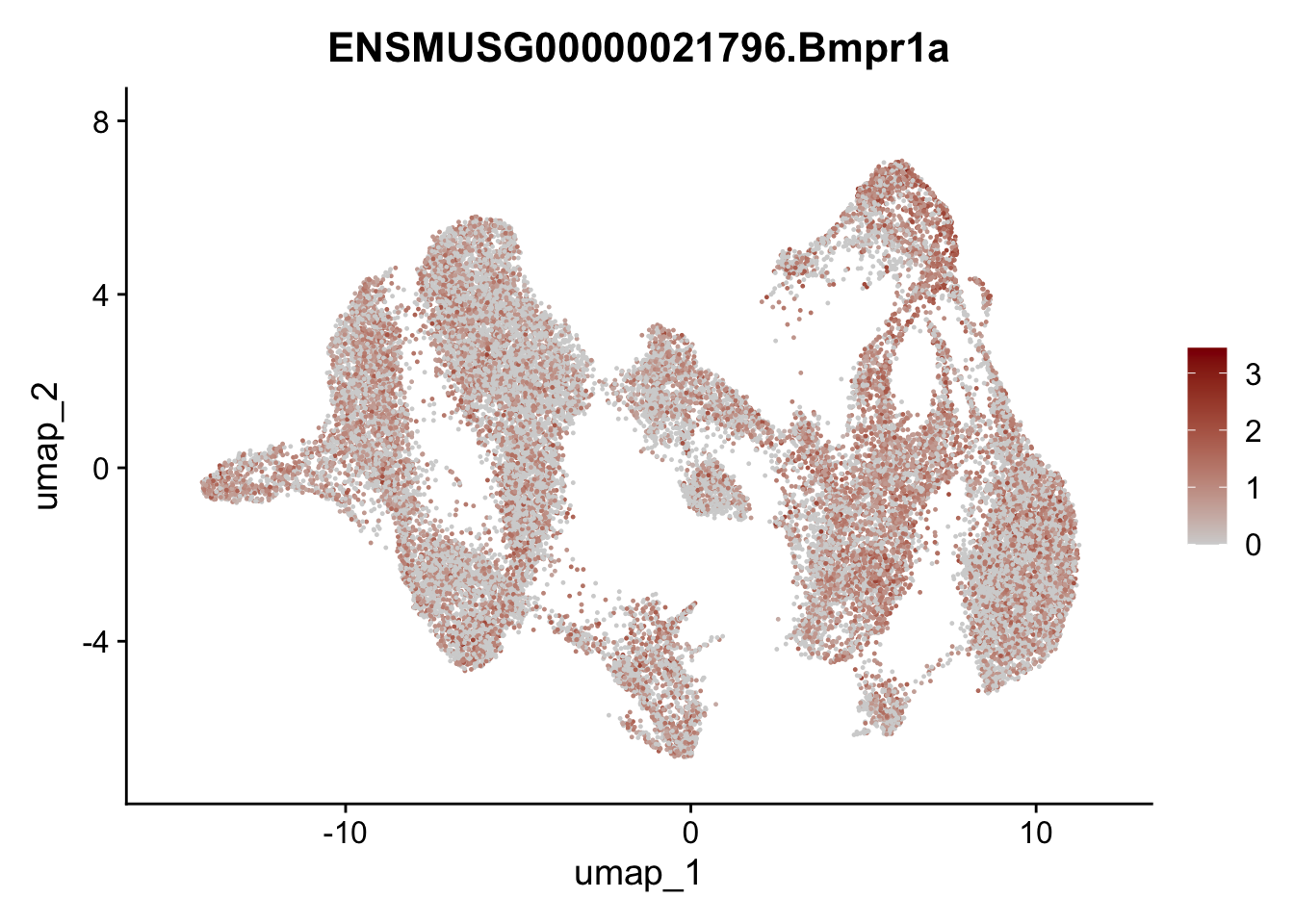
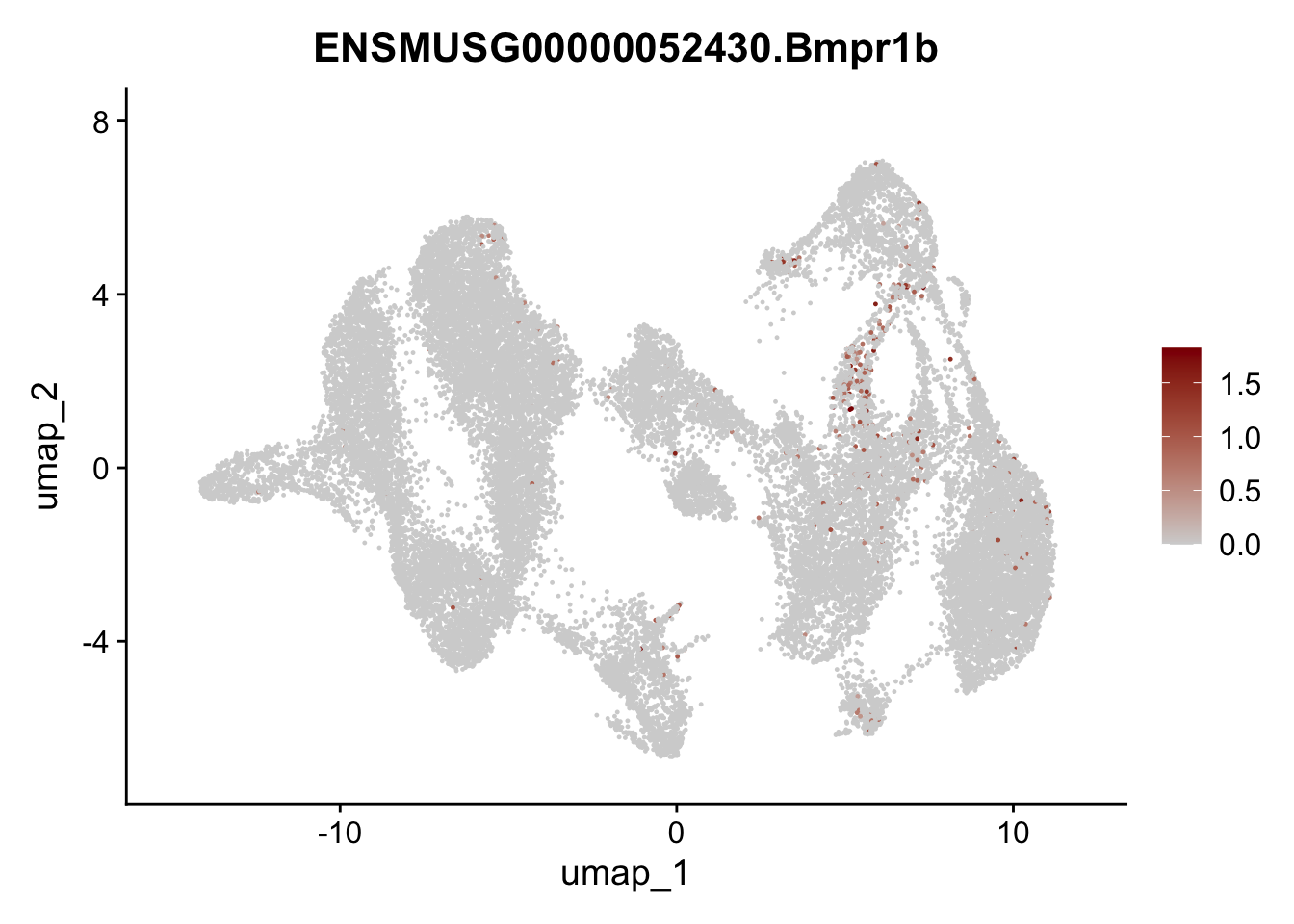
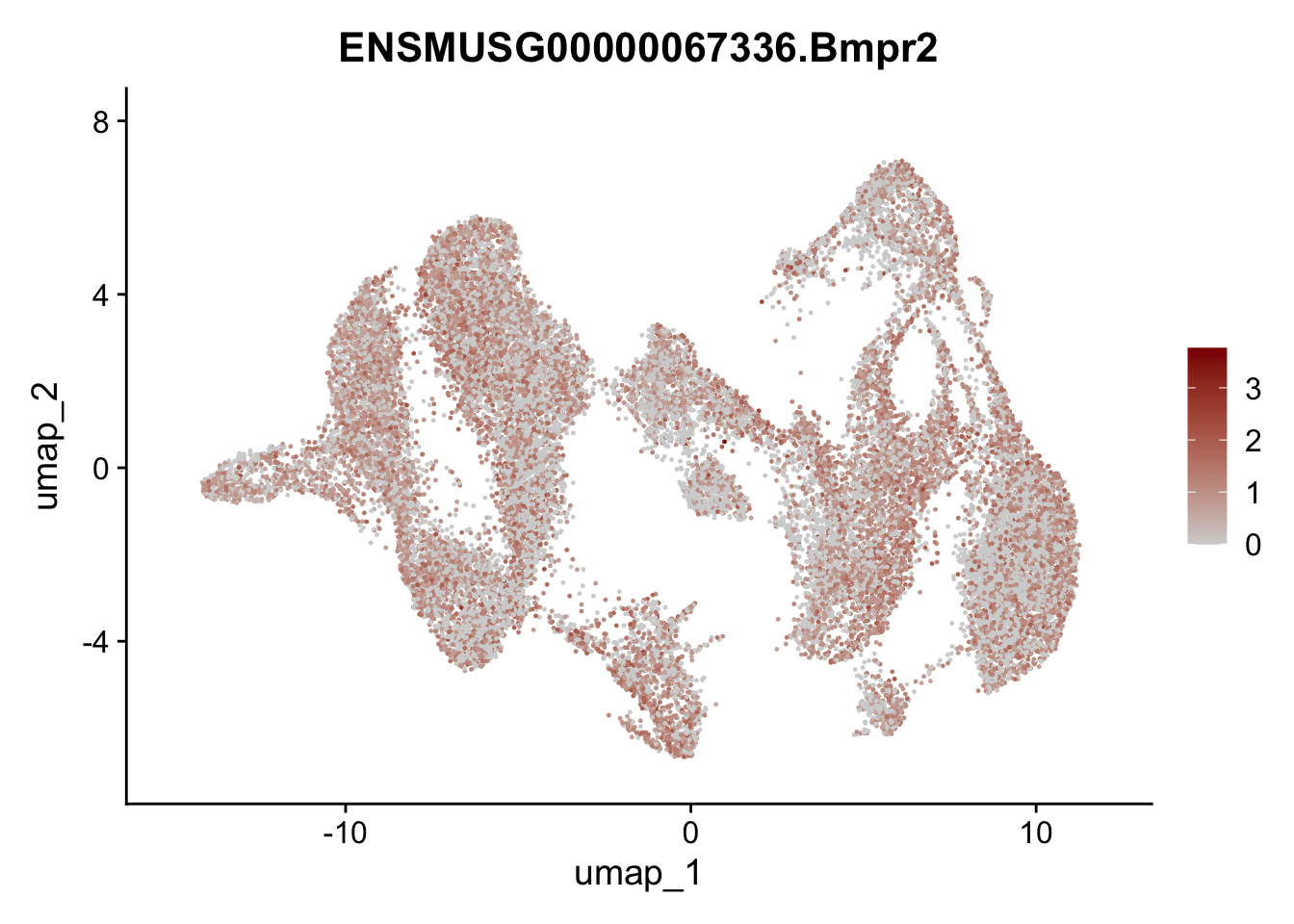
## save object
saveRDS(seuratmLNf3, file=paste0(basedir,"/data/LNmLToRev_EYFP_mLNf3_seurat.rds"))slingshot
sce <- as.SingleCellExperiment(seuratmLNf3)
sce <- slingshot(sce, clusterLabels = 'RNA_snn_res.0.4', reducedDim = 'UMAP',
start.clus = "1", end.clus = c("12", "3", "4", "7", "6"),
dist.method="simple", extend = 'n', stretch=0)load slingshot sce object mLN EYFP+
fileNam <- paste0(basedir, "/data/LNmLToRev_EYFP_mLNf3_slingshot_v2_sce.rds")
scemLNf3v2 <- readRDS(fileNam)clustDat <- data.frame(clustCol=colPal) %>% rownames_to_column(., "cluster")
timepointDat <- data.frame(ageCol=coltimepoint) %>% rownames_to_column(., "timepoint")
colDat <- data.frame(cluster=scemLNf3v2$RNA_snn_res.0.4) %>%
mutate(timepoint=scemLNf3v2$timepoint) %>% left_join(., clustDat, by="cluster") %>%
left_join(., timepointDat, by="timepoint")plot(reducedDims(scemLNf3v2)$UMAP, col = colDat$clustCol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, type = 'lineages', col = 'black')
plot(reducedDims(scemLNf3v2)$UMAP, col = colDat$ageCol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, type = 'lineages', col = 'black')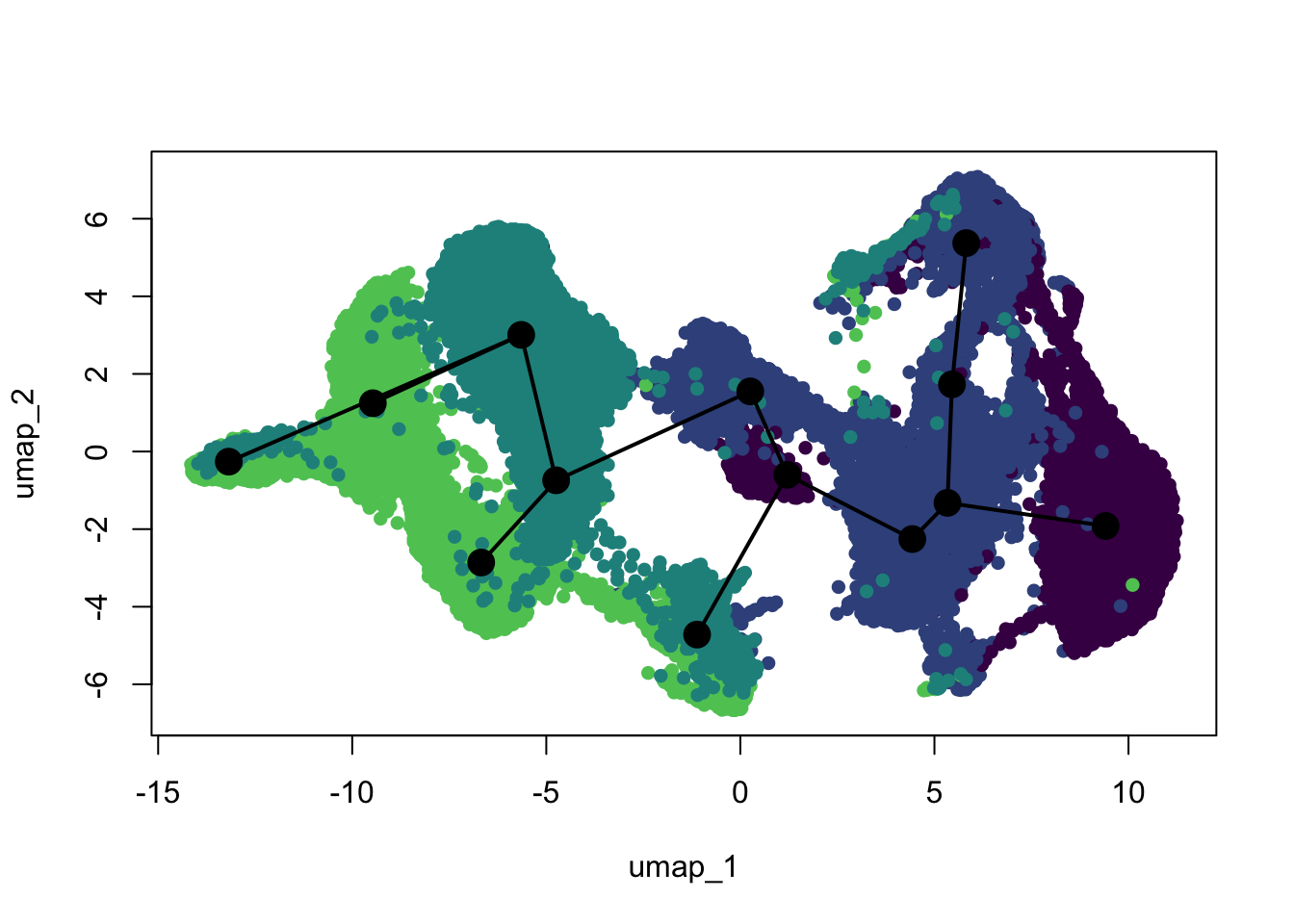
plot(reducedDims(scemLNf3v2)$UMAP, col = colDat$clustCol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, col='black')
plot(reducedDims(scemLNf3v2)$UMAP, col = colDat$ageCol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, col='black')
saveRDS(sce, file=paste0(basedir,"/data/LNmLToRev_EYFP_mLNf3_slingshot_v2_sce.rds"))summary(scemLNf3v2$slingPseudotime_1) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.000 2.715 10.207 10.639 17.741 25.951 9243 summary(scemLNf3v2$slingPseudotime_2) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.000 2.943 10.456 10.720 17.883 22.520 8873 summary(scemLNf3v2$slingPseudotime_3) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.000 1.959 6.353 8.476 15.403 20.789 12868 summary(scemLNf3v2$slingPseudotime_4) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.000 1.320 4.750 5.326 7.410 15.908 16994 summary(scemLNf3v2$slingPseudotime_5) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.000 1.315 5.106 5.355 8.250 14.665 17104 colors <- colorRampPalette(rev(brewer.pal(11,'Spectral')))(100)
plotcol <- colors[cut(slingAvgPseudotime(SlingshotDataSet(scemLNf3v2)), breaks=100)]
plot(reducedDims(scemLNf3v2)$UMAP, col = plotcol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, col='black')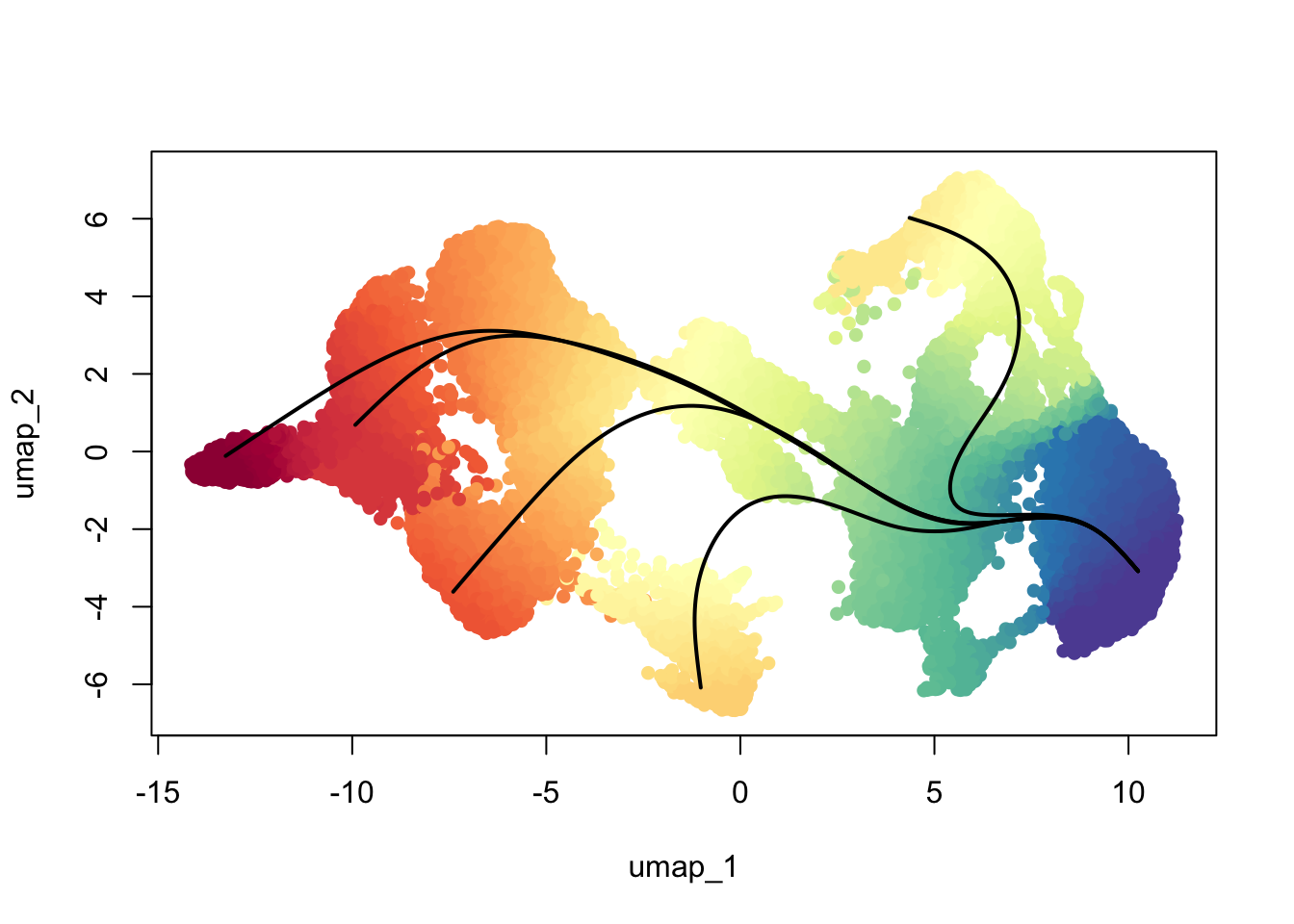
colors <- colorRampPalette(brewer.pal(11,'YlOrRd'))(100)
plotcol <- colors[cut(slingAvgPseudotime(SlingshotDataSet(scemLNf3v2)), breaks=100)]
plot(reducedDims(scemLNf3v2)$UMAP, col = plotcol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, col='black')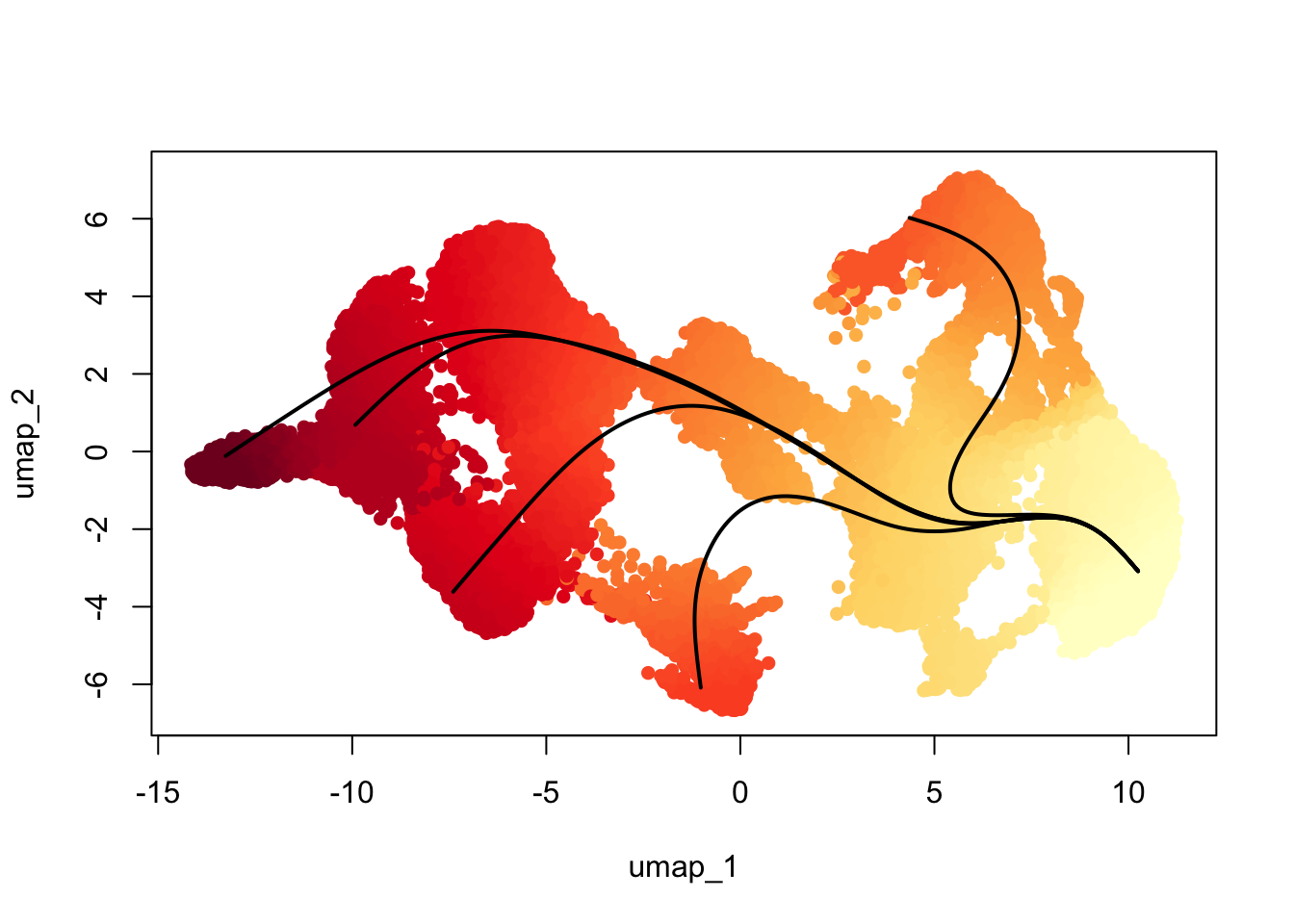
colors <- colorRampPalette(brewer.pal(11,'YlGnBu'))(100)
plotcol <- colors[cut(slingAvgPseudotime(SlingshotDataSet(scemLNf3v2)), breaks=100)]
plot(reducedDims(scemLNf3v2)$UMAP, col = plotcol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, col='black')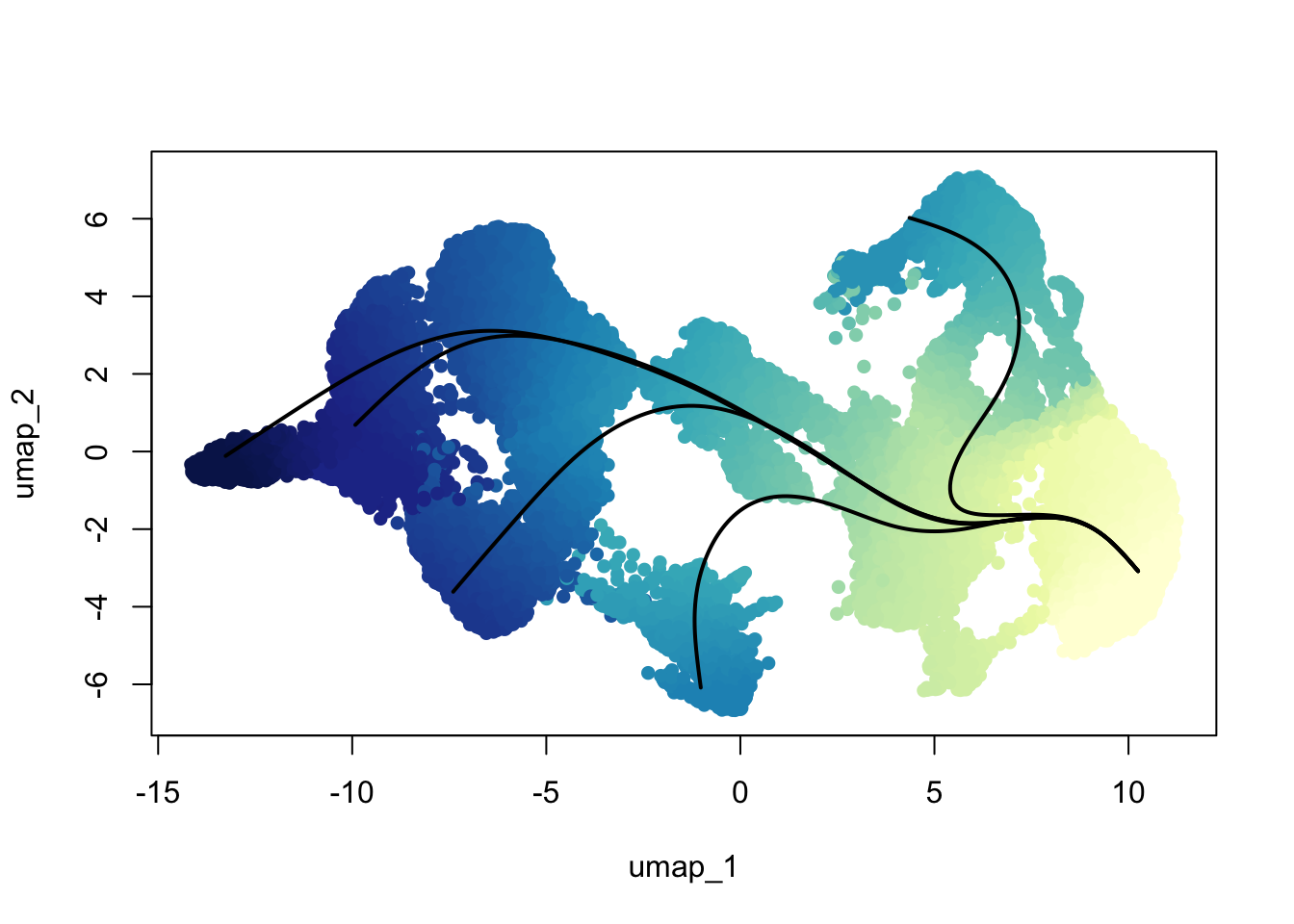
colors <- colorRampPalette(brewer.pal(11,'PuOr')[-6])(100)
plotcol <- colors[cut(slingAvgPseudotime(SlingshotDataSet(scemLNf3v2)), breaks=100)]
plot(reducedDims(scemLNf3v2)$UMAP, col = plotcol, pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=2, col='black')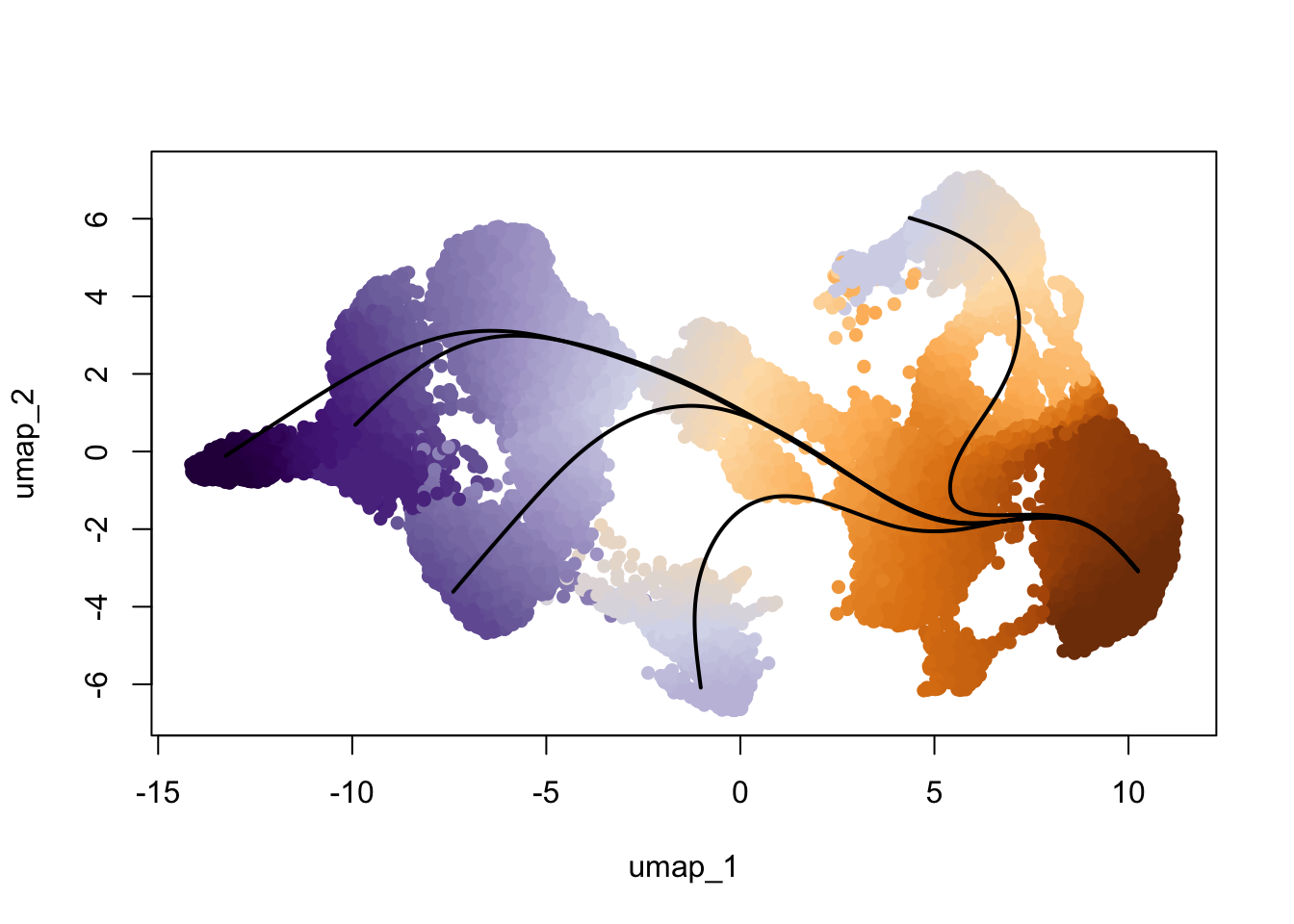
### color lineages
colLin <- c("#42a071","#900C3F","#424671","#e3953d","#b6856e")
names(colLin) <- c("1", "2", "3", "4", "5")
plot(reducedDims(scemLNf3v2)$UMAP, col = "#bfbcbd", pch=16, asp = 1)
lines(SlingshotDataSet(scemLNf3v2), lwd=4, col=colLin)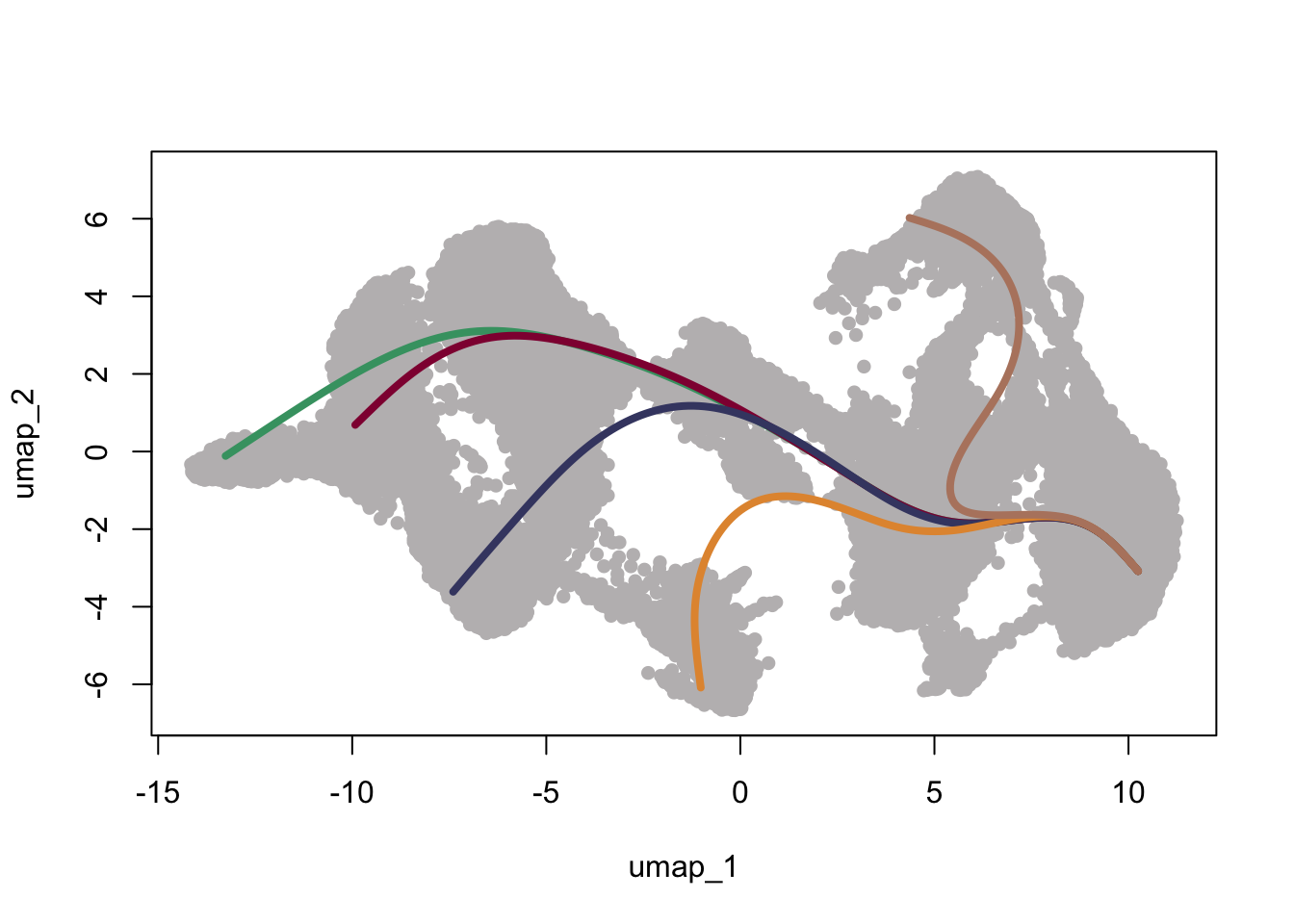
session info
date()[1] "Tue Jul 15 11:26:41 2025"sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-apple-darwin20
Running under: macOS Ventura 13.7.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Zurich
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] here_1.0.1 RColorBrewer_1.1-3 slingshot_2.12.0
[4] TrajectoryUtils_1.12.0 princurve_2.1.6 NCmisc_1.2.0
[7] VennDiagram_1.7.3 futile.logger_1.4.3 ggupset_0.4.1
[10] gridExtra_2.3 DOSE_3.30.5 enrichplot_1.24.4
[13] msigdbr_24.1.0 org.Hs.eg.db_3.19.1 AnnotationDbi_1.66.0
[16] clusterProfiler_4.12.6 multtest_2.60.0 metap_1.12
[19] scater_1.32.1 scuttle_1.14.0 destiny_3.18.0
[22] circlize_0.4.16 muscat_1.18.0 viridis_0.6.5
[25] viridisLite_0.4.2 lubridate_1.9.4 forcats_1.0.0
[28] stringr_1.5.1 purrr_1.0.4 readr_2.1.5
[31] tidyr_1.3.1 tibble_3.2.1 tidyverse_2.0.0
[34] dplyr_1.1.4 SingleCellExperiment_1.26.0 SummarizedExperiment_1.34.0
[37] Biobase_2.64.0 GenomicRanges_1.56.2 GenomeInfoDb_1.40.1
[40] IRanges_2.38.1 S4Vectors_0.42.1 BiocGenerics_0.50.0
[43] MatrixGenerics_1.16.0 matrixStats_1.5.0 pheatmap_1.0.13
[46] ggpubr_0.6.0 ggplot2_3.5.2 Seurat_5.3.0
[49] SeuratObject_5.1.0 sp_2.2-0 runSeurat3_0.1.0
[52] ExploreSCdataSeurat3_0.1.0
loaded via a namespace (and not attached):
[1] igraph_2.1.4 ica_1.0-3 plotly_4.10.4
[4] Formula_1.2-5 zlibbioc_1.50.0 tidyselect_1.2.1
[7] bit_4.6.0 doParallel_1.0.17 clue_0.3-66
[10] lattice_0.22-7 rjson_0.2.23 blob_1.2.4
[13] S4Arrays_1.4.1 pbkrtest_0.5.4 parallel_4.4.0
[16] png_0.1-8 plotrix_3.8-4 cli_3.6.5
[19] ggplotify_0.1.2 goftest_1.2-3 VIM_6.2.2
[22] variancePartition_1.34.0 BiocNeighbors_1.22.0 shadowtext_0.1.4
[25] uwot_0.2.3 curl_6.2.3 tidytree_0.4.6
[28] mime_0.13 evaluate_1.0.3 ComplexHeatmap_2.20.0
[31] stringi_1.8.7 backports_1.5.0 lmerTest_3.1-3
[34] qqconf_1.3.2 httpuv_1.6.16 magrittr_2.0.3
[37] rappdirs_0.3.3 splines_4.4.0 ggraph_2.2.1
[40] sctransform_0.4.2 ggbeeswarm_0.7.2 DBI_1.2.3
[43] jquerylib_0.1.4 smoother_1.3 withr_3.0.2
[46] git2r_0.36.2 corpcor_1.6.10 reformulas_0.4.1
[49] class_7.3-23 rprojroot_2.0.4 lmtest_0.9-40
[52] tidygraph_1.3.1 formatR_1.14 colourpicker_1.3.0
[55] htmlwidgets_1.6.4 fs_1.6.6 ggrepel_0.9.6
[58] labeling_0.4.3 fANCOVA_0.6-1 SparseArray_1.4.8
[61] DESeq2_1.44.0 ranger_0.17.0 DEoptimR_1.1-3-1
[64] reticulate_1.42.0 hexbin_1.28.5 zoo_1.8-14
[67] XVector_0.44.0 knitr_1.50 ggplot.multistats_1.0.1
[70] UCSC.utils_1.0.0 RhpcBLASctl_0.23-42 timechange_0.3.0
[73] foreach_1.5.2 patchwork_1.3.0 caTools_1.18.3
[76] data.table_1.17.4 ggtree_3.12.0 R.oo_1.27.1
[79] RSpectra_0.16-2 irlba_2.3.5.1 fastDummies_1.7.5
[82] gridGraphics_0.5-1 lazyeval_0.2.2 yaml_2.3.10
[85] survival_3.8-3 scattermore_1.2 crayon_1.5.3
[88] RcppAnnoy_0.0.22 progressr_0.15.1 tweenr_2.0.3
[91] later_1.4.2 ggridges_0.5.6 codetools_0.2-20
[94] GlobalOptions_0.1.2 aod_1.3.3 KEGGREST_1.44.1
[97] Rtsne_0.17 shape_1.4.6.1 limma_3.60.6
[100] pkgconfig_2.0.3 TMB_1.9.17 spatstat.univar_3.1-3
[103] mathjaxr_1.8-0 EnvStats_3.1.0 aplot_0.2.5
[106] scatterplot3d_0.3-44 ape_5.8-1 spatstat.sparse_3.1-0
[109] xtable_1.8-4 car_3.1-3 plyr_1.8.9
[112] httr_1.4.7 rbibutils_2.3 tools_4.4.0
[115] globals_0.18.0 beeswarm_0.4.0 broom_1.0.8
[118] nlme_3.1-168 lambda.r_1.2.4 assertthat_0.2.1
[121] lme4_1.1-37 digest_0.6.37 numDeriv_2016.8-1.1
[124] Matrix_1.7-3 farver_2.1.2 tzdb_0.5.0
[127] remaCor_0.0.18 reshape2_1.4.4 yulab.utils_0.2.0
[130] glue_1.8.0 cachem_1.1.0 polyclip_1.10-7
[133] generics_0.1.4 Biostrings_2.72.1 mvtnorm_1.3-3
[136] parallelly_1.45.0 mnormt_2.1.1 statmod_1.5.0
[139] RcppHNSW_0.6.0 ScaledMatrix_1.12.0 carData_3.0-5
[142] minqa_1.2.8 pbapply_1.7-2 httr2_1.1.2
[145] spam_2.11-1 gson_0.1.0 graphlayouts_1.2.2
[148] gtools_3.9.5 ggsignif_0.6.4 RcppEigen_0.3.4.0.2
[151] shiny_1.10.0 GenomeInfoDbData_1.2.12 glmmTMB_1.1.11
[154] R.utils_2.13.0 memoise_2.0.1 rmarkdown_2.29
[157] scales_1.4.0 R.methodsS3_1.8.2 future_1.58.0
[160] RANN_2.6.2 Cairo_1.6-2 spatstat.data_3.1-6
[163] rstudioapi_0.17.1 cluster_2.1.8.1 mutoss_0.1-13
[166] spatstat.utils_3.1-4 hms_1.1.3 fitdistrplus_1.2-2
[169] cowplot_1.1.3 colorspace_2.1-1 rlang_1.1.6
[172] DelayedMatrixStats_1.26.0 sparseMatrixStats_1.16.0 xts_0.14.1
[175] dotCall64_1.2 shinydashboard_0.7.3 ggforce_0.4.2
[178] laeken_0.5.3 mgcv_1.9-3 xfun_0.52
[181] e1071_1.7-16 TH.data_1.1-3 iterators_1.0.14
[184] abind_1.4-8 GOSemSim_2.30.2 treeio_1.28.0
[187] futile.options_1.0.1 bitops_1.0-9 Rdpack_2.6.4
[190] promises_1.3.3 scatterpie_0.2.4 RSQLite_2.4.0
[193] qvalue_2.36.0 sandwich_3.1-1 fgsea_1.30.0
[196] DelayedArray_0.30.1 proxy_0.4-27 GO.db_3.19.1
[199] compiler_4.4.0 prettyunits_1.2.0 boot_1.3-31
[202] beachmat_2.20.0 listenv_0.9.1 Rcpp_1.0.14
[205] edgeR_4.2.2 workflowr_1.7.1 BiocSingular_1.20.0
[208] tensor_1.5 MASS_7.3-65 progress_1.2.3
[211] BiocParallel_1.38.0 babelgene_22.9 spatstat.random_3.4-1
[214] R6_2.6.1 fastmap_1.2.0 multcomp_1.4-28
[217] fastmatch_1.1-6 rstatix_0.7.2 vipor_0.4.7
[220] TTR_0.24.4 ROCR_1.0-11 TFisher_0.2.0
[223] rsvd_1.0.5 vcd_1.4-13 nnet_7.3-20
[226] gtable_0.3.6 KernSmooth_2.23-26 miniUI_0.1.2
[229] deldir_2.0-4 htmltools_0.5.8.1 ggthemes_5.1.0
[232] bit64_4.6.0-1 spatstat.explore_3.4-3 lifecycle_1.0.4
[235] blme_1.0-6 nloptr_2.2.1 sass_0.4.10
[238] vctrs_0.6.5 robustbase_0.99-4-1 spatstat.geom_3.4-1
[241] sn_2.1.1 ggfun_0.1.8 future.apply_1.11.3
[244] bslib_0.9.0 pillar_1.10.2 gplots_3.2.0
[247] pcaMethods_1.96.0 locfit_1.5-9.12 jsonlite_2.0.0
[250] GetoptLong_1.0.5
sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-apple-darwin20
Running under: macOS Ventura 13.7.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Zurich
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] here_1.0.1 RColorBrewer_1.1-3 slingshot_2.12.0
[4] TrajectoryUtils_1.12.0 princurve_2.1.6 NCmisc_1.2.0
[7] VennDiagram_1.7.3 futile.logger_1.4.3 ggupset_0.4.1
[10] gridExtra_2.3 DOSE_3.30.5 enrichplot_1.24.4
[13] msigdbr_24.1.0 org.Hs.eg.db_3.19.1 AnnotationDbi_1.66.0
[16] clusterProfiler_4.12.6 multtest_2.60.0 metap_1.12
[19] scater_1.32.1 scuttle_1.14.0 destiny_3.18.0
[22] circlize_0.4.16 muscat_1.18.0 viridis_0.6.5
[25] viridisLite_0.4.2 lubridate_1.9.4 forcats_1.0.0
[28] stringr_1.5.1 purrr_1.0.4 readr_2.1.5
[31] tidyr_1.3.1 tibble_3.2.1 tidyverse_2.0.0
[34] dplyr_1.1.4 SingleCellExperiment_1.26.0 SummarizedExperiment_1.34.0
[37] Biobase_2.64.0 GenomicRanges_1.56.2 GenomeInfoDb_1.40.1
[40] IRanges_2.38.1 S4Vectors_0.42.1 BiocGenerics_0.50.0
[43] MatrixGenerics_1.16.0 matrixStats_1.5.0 pheatmap_1.0.13
[46] ggpubr_0.6.0 ggplot2_3.5.2 Seurat_5.3.0
[49] SeuratObject_5.1.0 sp_2.2-0 runSeurat3_0.1.0
[52] ExploreSCdataSeurat3_0.1.0
loaded via a namespace (and not attached):
[1] igraph_2.1.4 ica_1.0-3 plotly_4.10.4
[4] Formula_1.2-5 zlibbioc_1.50.0 tidyselect_1.2.1
[7] bit_4.6.0 doParallel_1.0.17 clue_0.3-66
[10] lattice_0.22-7 rjson_0.2.23 blob_1.2.4
[13] S4Arrays_1.4.1 pbkrtest_0.5.4 parallel_4.4.0
[16] png_0.1-8 plotrix_3.8-4 cli_3.6.5
[19] ggplotify_0.1.2 goftest_1.2-3 VIM_6.2.2
[22] variancePartition_1.34.0 BiocNeighbors_1.22.0 shadowtext_0.1.4
[25] uwot_0.2.3 curl_6.2.3 tidytree_0.4.6
[28] mime_0.13 evaluate_1.0.3 ComplexHeatmap_2.20.0
[31] stringi_1.8.7 backports_1.5.0 lmerTest_3.1-3
[34] qqconf_1.3.2 httpuv_1.6.16 magrittr_2.0.3
[37] rappdirs_0.3.3 splines_4.4.0 ggraph_2.2.1
[40] sctransform_0.4.2 ggbeeswarm_0.7.2 DBI_1.2.3
[43] jquerylib_0.1.4 smoother_1.3 withr_3.0.2
[46] git2r_0.36.2 corpcor_1.6.10 reformulas_0.4.1
[49] class_7.3-23 rprojroot_2.0.4 lmtest_0.9-40
[52] tidygraph_1.3.1 formatR_1.14 colourpicker_1.3.0
[55] htmlwidgets_1.6.4 fs_1.6.6 ggrepel_0.9.6
[58] labeling_0.4.3 fANCOVA_0.6-1 SparseArray_1.4.8
[61] DESeq2_1.44.0 ranger_0.17.0 DEoptimR_1.1-3-1
[64] reticulate_1.42.0 hexbin_1.28.5 zoo_1.8-14
[67] XVector_0.44.0 knitr_1.50 ggplot.multistats_1.0.1
[70] UCSC.utils_1.0.0 RhpcBLASctl_0.23-42 timechange_0.3.0
[73] foreach_1.5.2 patchwork_1.3.0 caTools_1.18.3
[76] data.table_1.17.4 ggtree_3.12.0 R.oo_1.27.1
[79] RSpectra_0.16-2 irlba_2.3.5.1 fastDummies_1.7.5
[82] gridGraphics_0.5-1 lazyeval_0.2.2 yaml_2.3.10
[85] survival_3.8-3 scattermore_1.2 crayon_1.5.3
[88] RcppAnnoy_0.0.22 progressr_0.15.1 tweenr_2.0.3
[91] later_1.4.2 ggridges_0.5.6 codetools_0.2-20
[94] GlobalOptions_0.1.2 aod_1.3.3 KEGGREST_1.44.1
[97] Rtsne_0.17 shape_1.4.6.1 limma_3.60.6
[100] pkgconfig_2.0.3 TMB_1.9.17 spatstat.univar_3.1-3
[103] mathjaxr_1.8-0 EnvStats_3.1.0 aplot_0.2.5
[106] scatterplot3d_0.3-44 ape_5.8-1 spatstat.sparse_3.1-0
[109] xtable_1.8-4 car_3.1-3 plyr_1.8.9
[112] httr_1.4.7 rbibutils_2.3 tools_4.4.0
[115] globals_0.18.0 beeswarm_0.4.0 broom_1.0.8
[118] nlme_3.1-168 lambda.r_1.2.4 assertthat_0.2.1
[121] lme4_1.1-37 digest_0.6.37 numDeriv_2016.8-1.1
[124] Matrix_1.7-3 farver_2.1.2 tzdb_0.5.0
[127] remaCor_0.0.18 reshape2_1.4.4 yulab.utils_0.2.0
[130] glue_1.8.0 cachem_1.1.0 polyclip_1.10-7
[133] generics_0.1.4 Biostrings_2.72.1 mvtnorm_1.3-3
[136] parallelly_1.45.0 mnormt_2.1.1 statmod_1.5.0
[139] RcppHNSW_0.6.0 ScaledMatrix_1.12.0 carData_3.0-5
[142] minqa_1.2.8 pbapply_1.7-2 httr2_1.1.2
[145] spam_2.11-1 gson_0.1.0 graphlayouts_1.2.2
[148] gtools_3.9.5 ggsignif_0.6.4 RcppEigen_0.3.4.0.2
[151] shiny_1.10.0 GenomeInfoDbData_1.2.12 glmmTMB_1.1.11
[154] R.utils_2.13.0 memoise_2.0.1 rmarkdown_2.29
[157] scales_1.4.0 R.methodsS3_1.8.2 future_1.58.0
[160] RANN_2.6.2 Cairo_1.6-2 spatstat.data_3.1-6
[163] rstudioapi_0.17.1 cluster_2.1.8.1 mutoss_0.1-13
[166] spatstat.utils_3.1-4 hms_1.1.3 fitdistrplus_1.2-2
[169] cowplot_1.1.3 colorspace_2.1-1 rlang_1.1.6
[172] DelayedMatrixStats_1.26.0 sparseMatrixStats_1.16.0 xts_0.14.1
[175] dotCall64_1.2 shinydashboard_0.7.3 ggforce_0.4.2
[178] laeken_0.5.3 mgcv_1.9-3 xfun_0.52
[181] e1071_1.7-16 TH.data_1.1-3 iterators_1.0.14
[184] abind_1.4-8 GOSemSim_2.30.2 treeio_1.28.0
[187] futile.options_1.0.1 bitops_1.0-9 Rdpack_2.6.4
[190] promises_1.3.3 scatterpie_0.2.4 RSQLite_2.4.0
[193] qvalue_2.36.0 sandwich_3.1-1 fgsea_1.30.0
[196] DelayedArray_0.30.1 proxy_0.4-27 GO.db_3.19.1
[199] compiler_4.4.0 prettyunits_1.2.0 boot_1.3-31
[202] beachmat_2.20.0 listenv_0.9.1 Rcpp_1.0.14
[205] edgeR_4.2.2 workflowr_1.7.1 BiocSingular_1.20.0
[208] tensor_1.5 MASS_7.3-65 progress_1.2.3
[211] BiocParallel_1.38.0 babelgene_22.9 spatstat.random_3.4-1
[214] R6_2.6.1 fastmap_1.2.0 multcomp_1.4-28
[217] fastmatch_1.1-6 rstatix_0.7.2 vipor_0.4.7
[220] TTR_0.24.4 ROCR_1.0-11 TFisher_0.2.0
[223] rsvd_1.0.5 vcd_1.4-13 nnet_7.3-20
[226] gtable_0.3.6 KernSmooth_2.23-26 miniUI_0.1.2
[229] deldir_2.0-4 htmltools_0.5.8.1 ggthemes_5.1.0
[232] bit64_4.6.0-1 spatstat.explore_3.4-3 lifecycle_1.0.4
[235] blme_1.0-6 nloptr_2.2.1 sass_0.4.10
[238] vctrs_0.6.5 robustbase_0.99-4-1 spatstat.geom_3.4-1
[241] sn_2.1.1 ggfun_0.1.8 future.apply_1.11.3
[244] bslib_0.9.0 pillar_1.10.2 gplots_3.2.0
[247] pcaMethods_1.96.0 locfit_1.5-9.12 jsonlite_2.0.0
[250] GetoptLong_1.0.5